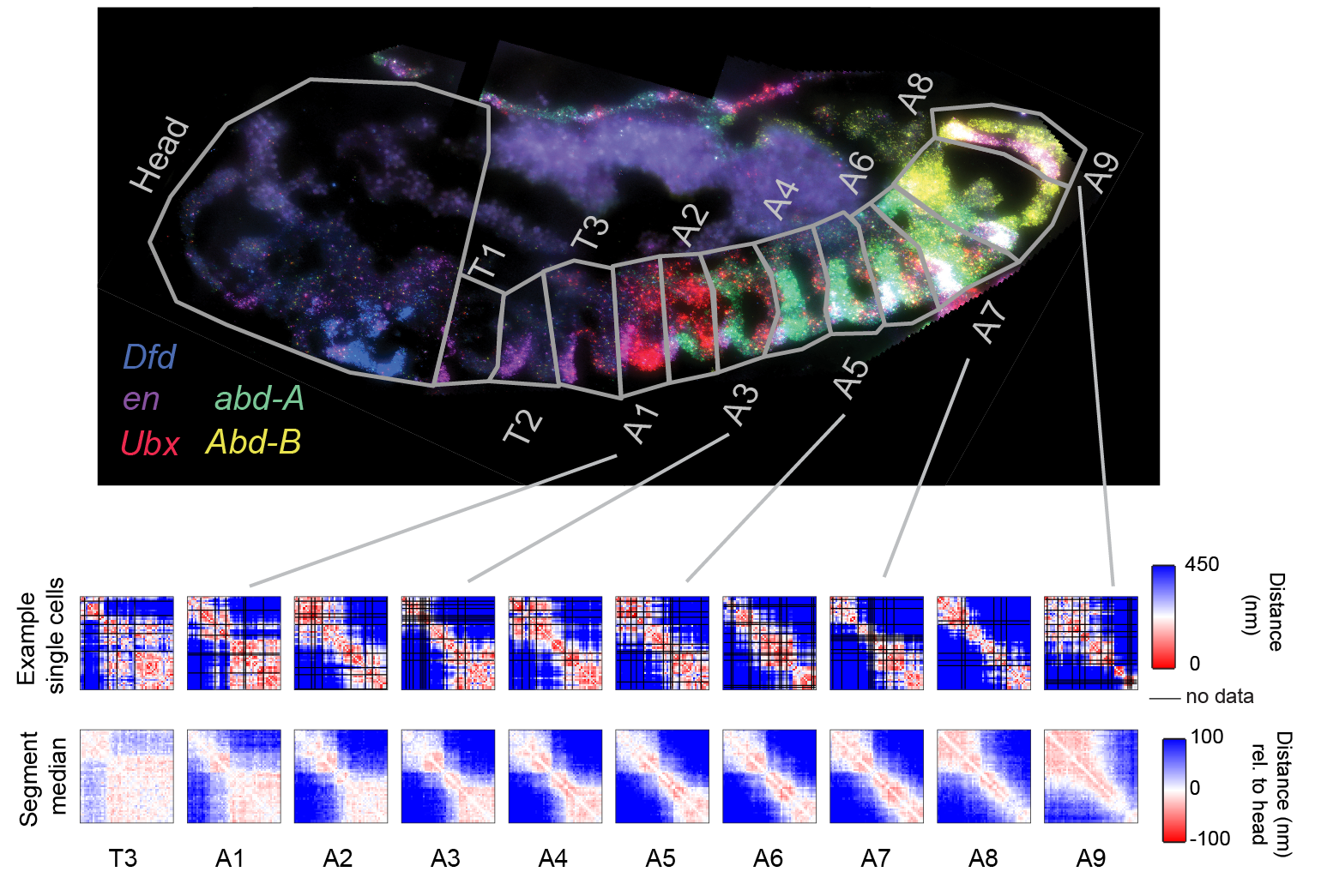
We are interested to learn to what extent changes in 3D genome folding facilitate changes in cell fate during development, using both fly and mouse development as a model system. In Drosophila, distinct levels of 3 posterior Hox genes specify the anterior-posterior cell identities in the 10 most posterior segments of the animal. The three genes are positioned next to one another in the genome, amidst a large gene desert densely packed with regulatory sequences. We have found that the genome is folded in a distinctive 3D structure in each of these 10 segments, altering the interaction frequencies among enhancers, promoters, and repressive elements to create 10 distinct expression states (Figure), (Mateo 2019).
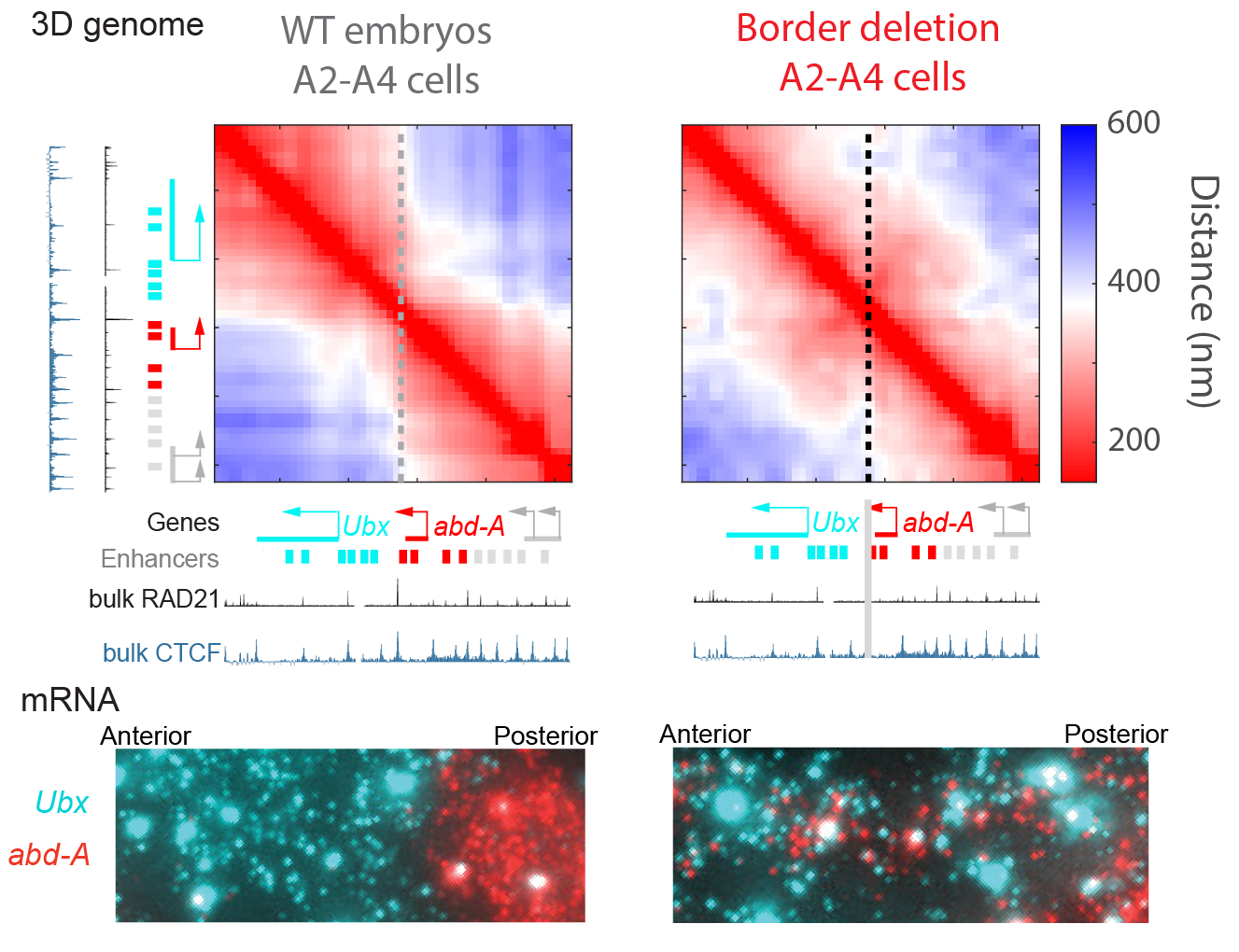
Deletion of enhancers in this locus results in misexpression of the Hox genes and corresponding homeotic transformations. The most famous of which, a triple deletion of enhancers that drive Ubx gene expression in the third-thoracic segment, converts the halteres into an extra set of wings, changing the fruit-fly to a dragon-fly appearance. However, genetic alterations that do not map to enhancers, repressors, or genes also have dramatic pheontypes in this locus.
We have found the strongest of these non-enhancer/gene mutations perturb the cell-type specific 3D structure, resulting in changes in cell fate (Mateo 2019). For example, deletion of a short region 3’ of the gene abd-A, which lies at a structural boundary (TAD boundary) we found results in a dramatic change to 3D structure, increasing proximity between both regulatory regions of both flanking genes and resulting in their misexpression. This deletion is embryonic lethal in homozygotes.
We continue to use hox loci to investigate roles of 3D genome structure in developmental patterning, taking advantage of their prominent roles in development, their complex regulatory landscapes, and their curious collinear organization in the genome.