In order to study the 3D DNA structures which regulate contacts between cis-regulatory elements (CREs) and their target transcribed elements (TEs), we are developing a super-resolution imaging approaches capable of resolving tens to hundreds of individual CREs, TEs, and intervening sequence in single cells of sectioned embryonic tissue and cultured cells at better than 20 nm resolution. We are applying this approach first to a selection of well-studied loci from Drosophila embryos and vertebrate cultured-cells, where the CREs have been mapped and the TEs they regulate identified through prior extensive genetic work. By observing the higher order structure of these loci in both wildtype and mutant conditions and in distinct cell-types that differ in TE expression at the locus, we aim to identify general principles that regulate physical interactions and shape expression. We plan to combine these experiments with simultaneous imaging of nascent RNA and mature transcripts in single cells, to determine which physical structures promote and which impede gene expression. We will also plan to use genetic manipulations and interspecies comparison of CREs to understand the sequence variation within and between species that result in phenotypic variation of key traits such as facial features and disease susceptibility.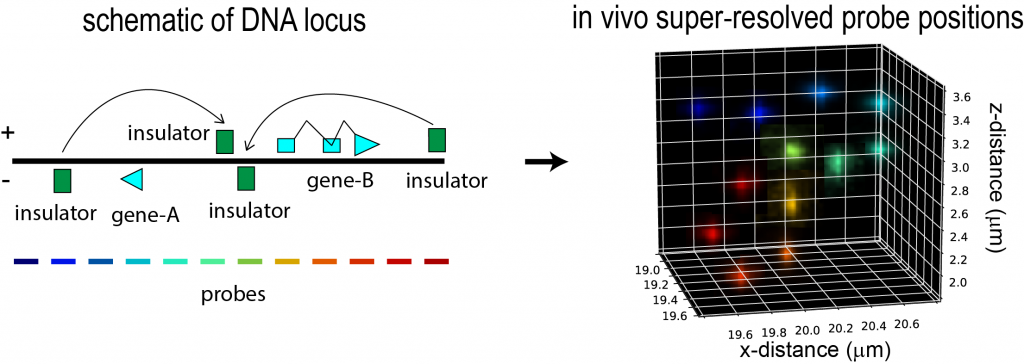
An illustration of the imaging approach to study CRE interactions: Left, a schematic of a locus containing two genes. Four cis-regulatory elements with insulator activity are labeled. The entire region has been tiled with 12 different colored probes, each overlapping different genetic elements (such as the promoters, coding sequences, and insulator elements) and the intervening sequence. Right, super-resolved localization of these 12 probe sets reveal the 3D organization of this locus in a single cell, allowing the identification of physical contacts and physically separated domains. The frequency of such contact can be determined by repeating this measurement across many cells. Each cell may be simultaneously labeled for expression of nascent RNAs from the two genes, and the structures correlated with level of expression.