Single molecule fluorescent in situ hybridization (FISH) is a powerful technique to label and count individual RNA molecules of a particular type inside a fixed cell or tissue. It allows us to study the organization of RNAs within a cell and the organization of cell-types (classified by RNA expression) within a complex tissue or environment. The ability to have precise molecular quantification enables us test biophysical models of gene regulation. Unfortunately the number of distinct species which can be studied simultaneously is limited by the number of spectrally distinct fluorophores / fluorophore combinations. While a postdoc in the Zhuang Group at Harvard University, Alistair co-developed a robust way to circumvent this limit using sequential addition and removal of fluors to a fixed sample, called MERFISH. Combined with error-correcting encoding schemes adapted from the field of digital communications, Multiplexed, Error Robust FISH (MERFISH) allows detection of 100s of species of RNA in intact cells and tissues, allowing systems level analysis of gene networks with molecular scale resolution.
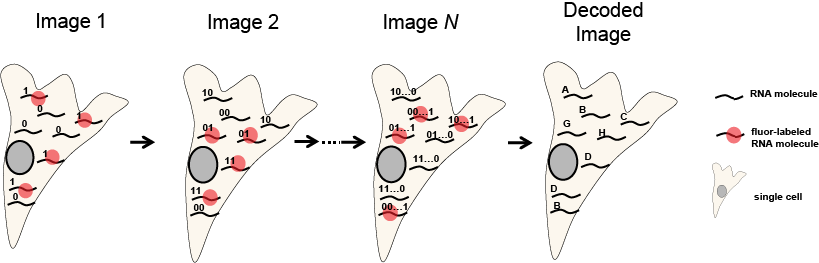
Multiple RNA species are labeled and imaged in the same color. The position of each label is recorded, the labels are then removed, new labels added, and the sample re-imaged. Over successive rounds of imaging a binary “word” is constructed at each position in space, with a “1” for each round in which a label was applied and detected and a “0” for each round in which that RNA was not labeled and thus not detected.
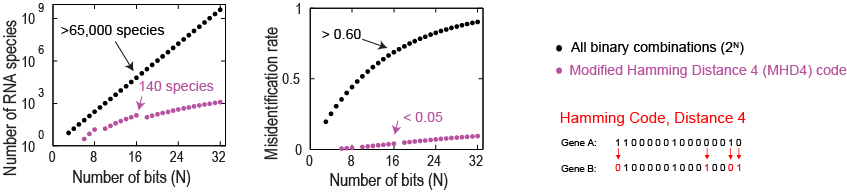
Choice of combinatoric scheme effects the number of labels and the error rate
The number of unique binary words and hence the number of RNA species grows exponentially with the number of imaging rounds (bits). However, the small error rates also grow exponentially. This growth in errors can be combated by using error detecting and correcting codes, which sacrifice the number of species. With this compromise, in 16 rounds we can still label 140 different RNA species. Using an error detecting code without error correction, we can encode 1001 species in 14 rounds.
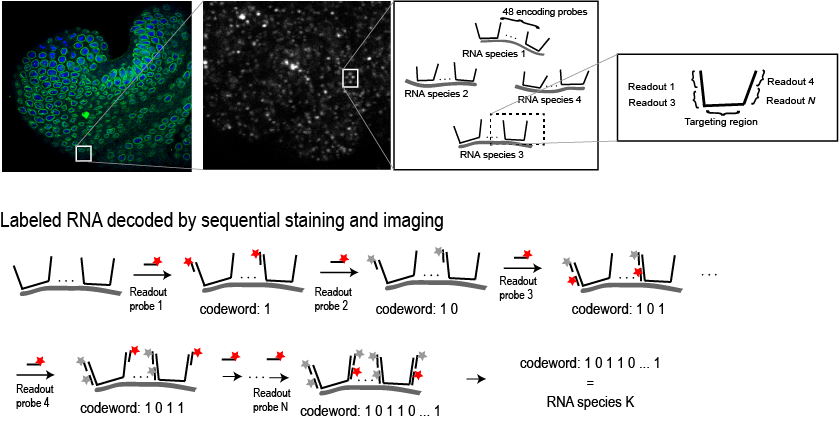
Encoding RNA with synthetic oligos and decoding by sequential imaging
Top image shows an embryo labeled with a MERFISH library. Blow ups show the probe design which allows sequential addition and removal of fluorescent labels in a rapid manor. Each readout probe takes only 15 minutes to hybridize.
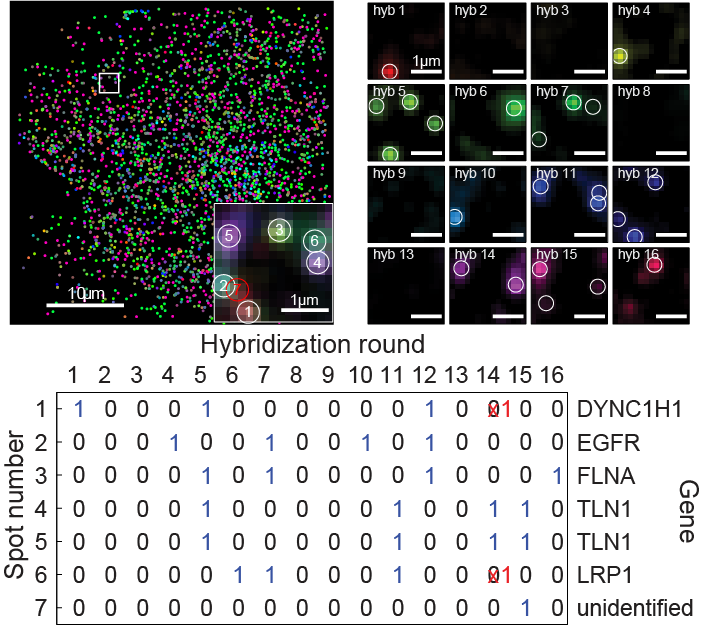
Example: 140 RNA species detected in a single human fibroblast
Image of a single human fibroblast with 140 different RNA species labeled in different colors. The panels to the right show raw images recorded in each round of hybridization and imaging for the blow-out indicated by the white square. The graph to the left records which round each RNA in the blow-region was detected in and decodes the corresponding binary word into its corresponding RNA species. Data shown here are taken from Chen, Boettiger, Moffitt, et al. 2015.
Validating the measurements with sequencing and smFISH
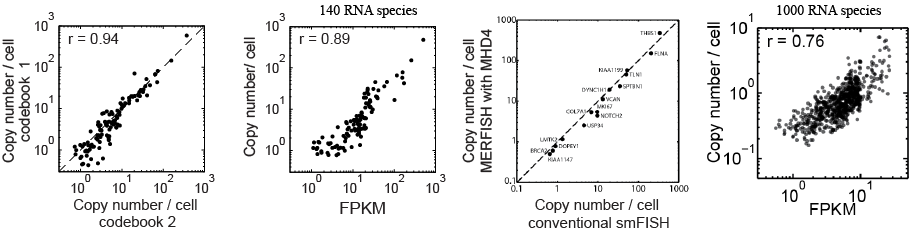
To test for bias in the encoding scheme, the same binary words were assigned to different RNA species in a second synthetic oligo library. This library was used to read out RNA copy number of all species and produced results indistinguishable from the first (left graph). The cells were also subjected to RNA-seq and the FPKM correlated to the measured copy number per cell by MERFISH, revealing a strong correlation (2nd graph). The average molecule counts per cell for a select set of genes was measured by smFISH, which correlated very strongly with the results by MERFISH (3rd graph). An alternate encoding with error detection only and no error correction, used to measure 1000 different RNAs, still produced high correlation with bulk sequencing, though the detection rate for individual RNAs was reduced (4th graph).