10:00 am – 12:45 am
Chromatin Modeling
initalization
- compressed random walk polymer maybe not best initial fractal globule state
- try using a sticky polymer, these have pretty good fractal structure all the way down.
- too sticky polymer settles down too slowly (even with local energy minimization).
- starting from this condensed state and letting the system relax rapidly back to the new target density is also bad
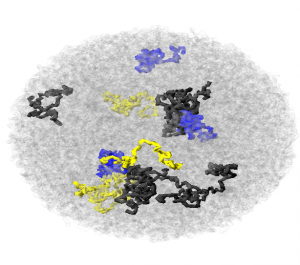
- in particular, the black chromatin rebounds faster than the yellow, and the yellow stays compressed.
Initial condition variation
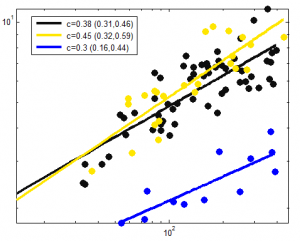
- starting on the same section of the same initial fractal globule has strong effects on behavior
- fractal globules having all scales of loops have dramatic variation in the size of any given domain. It seems to me insufficient attention has been given to this variability in the literature.
effects of Energy minimization
- energy minimization greatly helps compacting blue domains
- energy minimization possibly expands the black domains at the expense of the softer yellow domains.
- maybe if the yellows are sufficiently transparent they won’t get compressed?
- not sure if this true — maybe just an artifact of the fact my circ permute domains got all my domains mixed up.
Debugging
- okay, debugged new permute domains. hopefully this works in disrupting the dependence on initial condition.
- things looking a bit more promising. Need more averaging!