10:05 am – 8:00 pm
RNAi
Re-analyzed data
- issues with row/column labels confused analysis
- round 1 results:
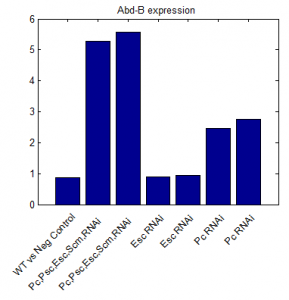
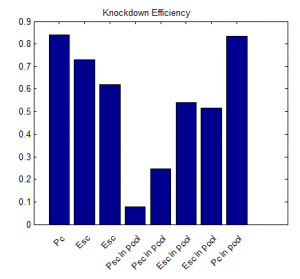
- repeated qPCR experiments with higher concentration cDNA
- round 2 results:
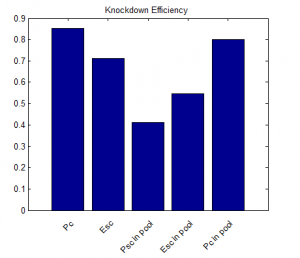
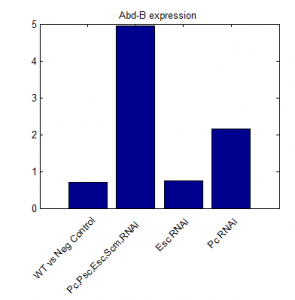
- looks promising!
- to do next
- proceed with staining some cells for Pc to validate at single cell level
- make more cDNA from isolated total RNA for deep sequencing.
New RNAi setup
- Plate 1
- row 1: Th 30 ug, Mock, Psc 60 uG
- row 2: Pc, Psc, Esc, Scm 30ug each, Ph-p + Ph-d, 40 ug each, Su(Z)2 30 ug
- Plate 2
- row 1: Psc 60 ug, Psc 60 ug, Psc 60 ug
- row 2: Ph 30 ug, mock, mock
Applications
- rewriting text of DF application
- working on figures for DF application
Conferences
- finished registration and payment for EMBO Nuclear structure meeting
MERFISH
- analyzing Guiping’s new data.
- 512×512 36 bits is running very slowly.
- its the upsampled CorrAlign command that kills the run speed.
- sped up CorrAlign
- doesn’t need to align every bit, just every hybe
- doesn’t need whole image, should work with just the center field of view (may have trouble, we’ll see).
- is this really supposed to be 36 bit data?
- where’s the codebook?
