9:10 am – 7:00 pm, 8:50pm – 1:00 am
MERFISH
- Analyzing MERFISH data, (9:10 am – 1:00 pm)
- see post
- MERFISH meeting (1:00 – 2:30 pm)
- Jeff’s talk (4:00 pm – 5:15 pm)
- rerunning analysis with nearest third of pixel alignment for drift (previously just nearest pixel, not good enough?)
- set to decode on non-upsampled images with lower thresholds. see if we get any real RNAs (I think the massive number of potential blanks still wash out this signal).
- should be able to run pipleline 2 on this data
- wrote new approach for L16, based on subtraction. (see notes)
To do
- check sorting of codebook and that final codes, names, and indices match.
RNAi
- discuss updates and plan of attack with BB (3:00 pm – 4:00 pm)
- out of dsRNA for more KD – amplifying more template (3x reaction in 3 strips of 6)
- probe order for PCR:
- Pc1,
- Pc2 (currently good),
- Ph-p,
- Ph-d,
- esc
- scm
- ran PCR cleanup (pooled triplicates, eluted in 20 uL ddH2O).
- setup new T7 reactions (20 uL DNTP buffer, 4 uL T7 mix, 2 uL RNAsin Plus) (10:30 pm)
- current PPhES KD on day 2
- analyze data: KD results from last batch
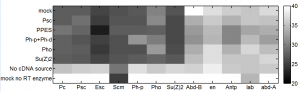
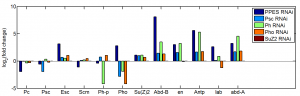
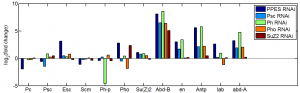
- ordered primers to GAPDH1, Act5C, and alphaTub84b from IDT (Chrome refuses to do this, redirect loop, ordered through firefox).
