9:30 am – 7:00 pm
Finalizing draft of response to reviewers
- polishing figures a bit (removing boxes, fixing axes etc)
- finish writing missing captions
- 9:30 am – 10:30 am
communication
- catching up on communication emails (10:30 – 11:15)
- coffee with NB
- reply to pairing hoedown invitation
- reply to meeting planning for CONTI retreat
Discussing deconvolution
- doesn’t shrink area estimated much
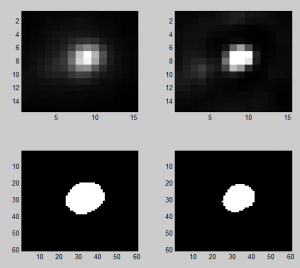
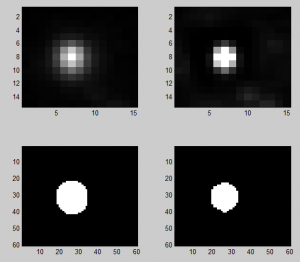
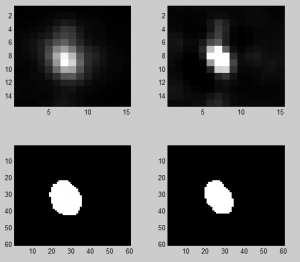
- maybe this is a problem? More iterations than the default do further tighten the PSF if you apply it to itself. In 10 it doesn’t shrink down to a point but it does in 60. Lots of iterations on sample data does not reduce it to a point, however the background noise goes a bit crazy.
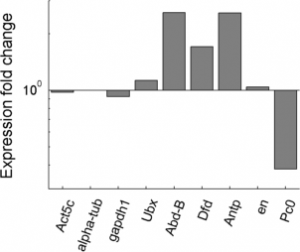
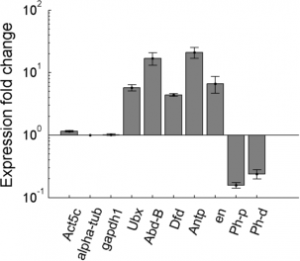
QPCR
- 3 Ph-KD with corresponding mocks (samples 1-6): test with 2 alternate ph-d primers (12 wells)
- 1 Pc-KD with corresponding mocks (samples 6 & 7): test with 2 Pc primers (4 wells)
- I had additional Pc-RNAi data from 7/3 with 2 replicates. Maybe I can find these samples again.
- sample order:
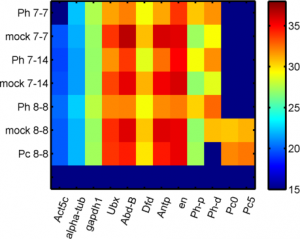
- row 1, col 1-6 ph-d primer set 1 (cDNA samples 1-6 from 8-14: ‘Ph 7-7′,’mock 7-7′,’Ph 7-14′,’mock 7-14′,’Ph 8-8′,’mock 8-8’)
- row 2, col 1-6 ph-d primer set 2 (cDNA samples 1-6 from 8-14: ‘Ph 7-7′,’mock 7-7′,’Ph 7-14′,’mock 7-14′,’Ph 8-8′,’mock 8-8’)
- row 7, 8 Pc primers 0 and 5 in Pc 8-8 KD and mock 8-8 (described in matlab script qPCR_RNAi_PcGs_150910b)l
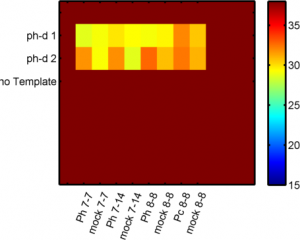
- data looks good, Ph-d primer 1 didn’t work but primer pair 2 looks decent.
RT for QPCR from Pc RNA
- samples 7-26 mock, 7-26 Pc v1, 7-26 Pc v2, 8/8? Pc KD (v2)
- RNAse H treated (ran a bit long, 50 min at 37C instead of 20 min)
- Ran DNA cleanup column, eluted in 30 uL
- diluted 20 uL into 40 uL 10 mM Tris pH 8.0
- setup new QPCR
- 4 cDNA samples, 11 primer pairs.
- 50x master mix: 625 uL 2x Phusion master + 312.5 ddH2O + 62.5 EvaGreen
-
- 3 uL cDNA + 2.5 uL 50 uM primer mix
- QPCR plate layout
- col primers: ‘Act5c’,’alpha-tub’,’gapdh1′,’Ubx’,’Abd-B’,’Dfd’,’Antp’,’en’,’Pc0′,’Pc1′,’Pc2′
- row cDNA ‘Pc KD 8/8’, ‘Pc v2 7/26′,’Pc v1 7/26′,’mock 7/26’
