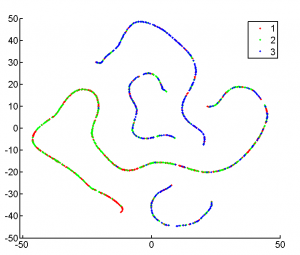
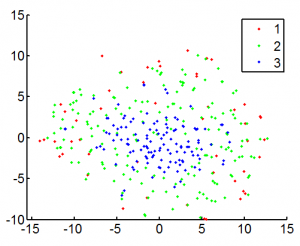
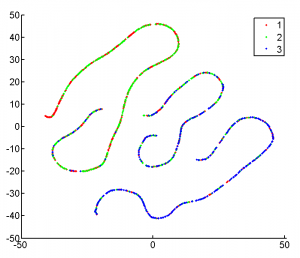
exploring tSNE (11 am -3:50 pm)
- looking at L7 data
- see the expected correlations (here in 3D)
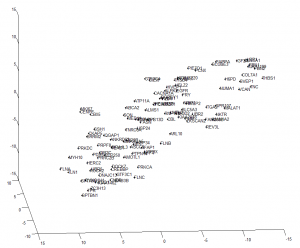
- can also see dataset biases (these go away on z-normalization).
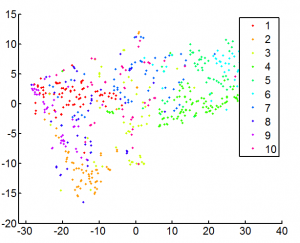
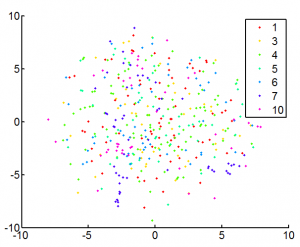
- 1000 gene data
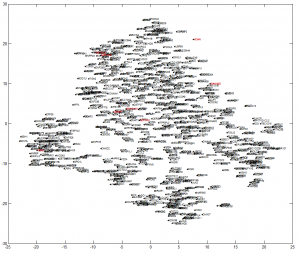
- not so good for chromatin structure yet
- algorithm cares about absolute orientation (forward and backward slanty 1s are different, 6s and 9s are different).
- could do some sort of global rotation to fix this, which would rotate all 1s in same direction
- still wouldn’t capture features likes lots of protrusions — its hierarchical clustering of absolute distances of the measurements.
- obviously our data descriptors like volume still have interesting affects on the data