9:30 am – 9:00 pm
ms review
- making notes on manuscript
applications
- rewriting intro / motivation paragraph.
new probes
- chr lib4 (MERFISH lib 5)
C01 primer_145 CCATCAGCCGCGACCCTATG chr3R:12691420-12706420__BXC
C02 primer_148 CTTTCTCGCAGGCGACTCGC chr3R:12706421-12721421__BXC
C03 primer_150 ACGTTAGGTGCCTCGCTGCG chr3R:12721422-12736422__BXC
C04 primer_152 CCCTCGGCGCAAAGTCTGTC chr3R:12736423-12751423__BXC
C05 primer_155 AATGCGGCGGAGTGACTTGC chr3R:12751424-12766424__BXC
C06 primer_157 CGATTTCGCGCCGCACTTTC chr3R:12766425-12781425__BXC
C07 primer_160 TTTGCTTGTGCGCCGGAAAG chr3R:12781426-12796426__BXC
C08 primer_162 GCTTCGCGTCTGCCGGAAGG chr3R:12796427-12810708__BXC
G01 primer_147 TAATACGACTCACTATAGGGCGACGGCTTTAGGGCCTCCG chr3R:12691420-12706420__BXC
G02 primer_149 TAATACGACTCACTATAGGGAAACGCGGGTGAACGCTCTG chr3R:12706421-12721421__BXC
G03 primer_151 TAATACGACTCACTATAGGGCCATATTCCGCGCATCTGGC chr3R:12721422-12736422__BXC
G04 primer_153 TAATACGACTCACTATAGGGCCGGACGAATCAGCCTTCC chr3R:12736423-12751423__BXC
G05 primer_156 TAATACGACTCACTATAGGGCGGACGGTCGGATTCTTGG chr3R:12751424-12766424__BXC
G06 primer_158 TAATACGACTCACTATAGGGCCGTCATCGCCGGAACTTTG chr3R:12766425-12781425__BXC
G07 primer_161 TAATACGACTCACTATAGGGCCCGTCTAGGCGCATGAACG chr3R:12781426-12796426__BXC
G08 primer_163 TAATACGACTCACTATAGGGTCAGATCGGCGCTCCGAATC chr3R:12796427-12810708__BXC
funny issues with amplification
- bubbles or something make very high starting levels of signal in QPCR in some chambers (5000 RU, baseline 2000 RU). This is as bright as the typical max
- as bubbles pop this drops down and then goes up again
- amplification is quite late.
- I think there are issues with this lib, test by gel
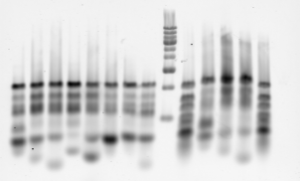
- try again after I submit my applications
- spec the primers (unfortunate — these were supposed to be diluted to fixed conc by IDT, maybe the need a good spin down?)
- go back to TSTORM-L5 source library — I think this dilution is no good.
- drop max on QPCR down to 28 cycles.
Cell culture and Knockdown
- moved samples to 28C for ~6 hours — RT is getting a bit cold maybe to keep up with KD, 28 is recommended culture condition.
New stains
- selected 3 coverslips
- incubated in 5% formaldhyde to dissolve sucrose and fix
- 10 min in .5% Triton
- rinse in PBS
- 2x SCC wash 10 min
- 50% formamide 2X SSC wash 10 min at 37 C
- add probe (1 uL probe + .7 uL S2-A647) denature at 78C upstairs (block is being unstable again :()
- heat block dropped from 78 to 73 (by both digital and analog thermometers) during 3 min denaturation of samples E22 and E23
- letting warm up again, denaturing sample E24
Some numbers on restriction enzymes
- AsiSI, an 8 cutter, 1 per megabase, 1000 target sites in the genome (probably too many)
- I-SceI, an 18 cutter (used by Mistelli) not in human genome.
- PI-PkoII 2CW7 Pyrococcus kodakaraensis KOD1 A 5′ CAGTACTACGGTTAC / 3′ GTCATGATGCCAATG
