10:00A – 10:50 P
- Bcd Gal4 x shRNA Pc2 and x Psc shRNA1 both still hatch larvae.
- Flip collection cages
- check mail for Perrimon Gal4 3x line – not yet arrived
- rerunning Pc-GFP controls with Colenso’s parameters (min height 750 = 80, min height 647 = 200, 561 = 100, 488 = 100). Compare image and single molecule photon characteristics. run started at noon
- in vitro transcription reactions:
- [spreadsheet 0AjSqkxgziU1YdFpWbVJoUmhSY2FPLXpsU3p4NkhQdEE 600 800 sheet=1]
- samples at -20C
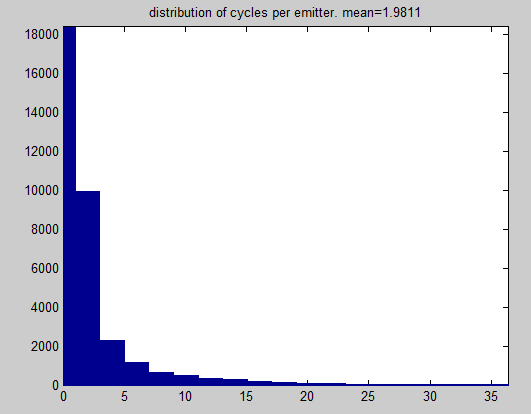
- writing code to do single molecule statistics analysis. Fixed bug created by using pixelated object centroids rather than CoM from raw localization data.
- Fixed bug to eliminate single localizations from analysis. Just do fitting on the molecules that blinked many times (i.e. lower-threshold of 5-10 localizations).
- Added z-resolution calculator
- Rewriting single molecule statistics script (previously part of scripts analyze_singlemol_stats.m and clust_onbin.m) into a separate molecule clustering function.
- Need to record goodness of fit, not just final choice gaussian parameters, for data to be meaningful.
- density histogram doesn’t place molecules into same sections as scatterplot x-y, need to troubleshoot with a small area.
- Fixed issue by specifying centers instead of number of bins.