9:45 A – 7:00P, 8:00P – 10:30P
Making summary images of SCM data.
Reconcentrating / Precipitating probe prior to nicking reaction.
- Combined into 2 tubes.
- Dissolving pellets at 37 O/N. (waiting for Nicking enzyme to continue).
- Nicking enzyme arrived
- Brian will help set up PAGE gels on Sunday. Need to run digest tomorrow
- Samples frozen at -20C.
DNA phenol extraction
- from frozen sim[D] Espl[D] flies.
- Preparing buffers for phenol extraction protocol
- Homogenization was supposed to happen in Buffer A. Maybe we should just get a tissue homoginizer.
- protocol pretty smooth, somewhat intenstive (this is going to be fun if we need to screen 100 recombinants this way).
- Don’t use air gun on pellets (fished pellet off desk. Will see later how it works.)
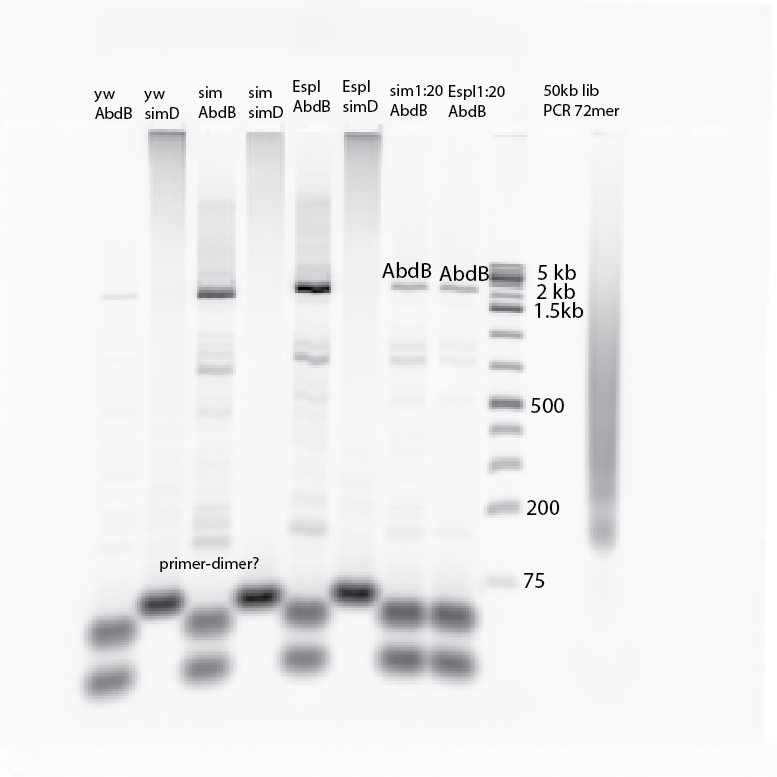
- Running PCR. Lanes: YW-AB, YW-SimD, sim-AB, sim-SimD, Espl-AB,Espl-SimD, sim_1:20-AB, Espl_1:20-AB
- All Abd-B amplifications worked. simD probes look like primer dimer (one band at bottom, larger than the AbdB primers, which seem to have two bands (not quite sure what these bands at the bottom are). Maybe over concentrated primer, don’t normally see it, though the Typhoon is somewhat more sensitive).
Fly work
- Collect virgins AM
- Collect virgins PM. Start crosses.
- Out of vials. Need to wait till next week to set up next round of crosses.
DNA FISH – (test cent probe co-localization with H3K9me2 )
- Fixing S2 cells.
- Went to sort flies, over-fixed cells, 35 min (goal 15)
- Incubating O/N on slides in newly built box at 37C with cent probe
- Test H3K9me2, H3K27me3, H3K27ac, Pc, Su(Z)12, Pho
Confocal test PcG colorflip embedded embryos. Dm0 singal very strong, nuclear signal very weak. Not good labeling. Should repeat and should do in separate channels.
