9:50 A – 7:45 P, 9:30 P – 12:15 A
Research update meeting
- preparing update on multi-color DFISH
- No data on BX-C vs Flank, I recall it didn’t work well but I thought I at least had an experimental record.
- diagram chromatin organization
- Bioinformatics for color classification
- Bioinformatic approach for selecting top target regions
- Bioinformatic approach for designing probe sets to respect sense and antisense, avoid repeats, internal genes, etc.
- Two nice very different sections of BX-C condensed vs. active. Overall most embryo dots look like the ‘active’ type. See post.
Feedback
- 4 colors, 3 length scales, 2 regions
- Domain mixing
Genomics
- UCSC whole genome data does include the heterochromatic regions and Y chromosome: http://genome.ucsc.edu/cgi-bin/hgTracks?db=dm3&chromInfoPage=
- OligoArray crashed on day 2 half way through chr2R. Should probably do the whole genome version of this chromosome by chromosome. (Should really probably run it on the cluster on 500 cores rather than 18). For the meantime can probably do on demand probe region by probe region analysis for 20 – 500 kb fragments.
Review
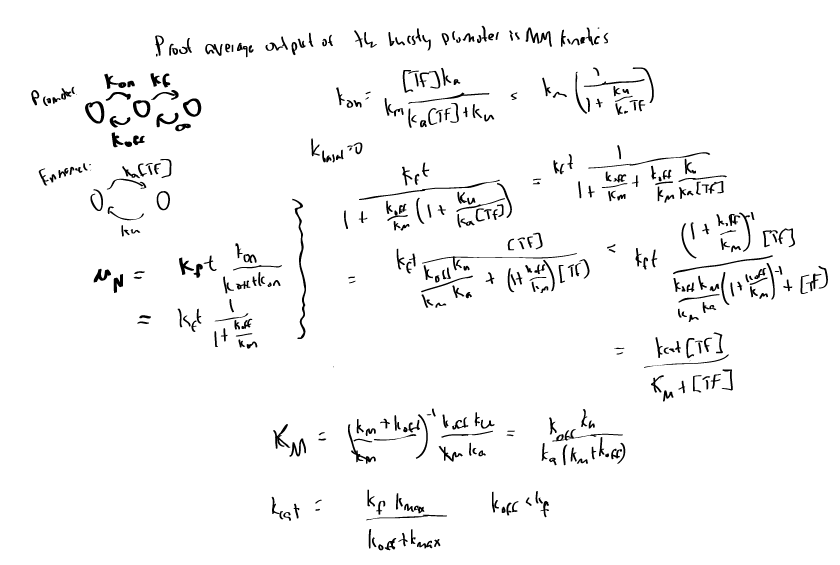
- working on transcription review — keep it simple, it’s a review not new stuff. Keep new stuff for the kinetics project!
- Finished first pass through Theory section, Derivation of common models section, and cooperativity subsection (latter still needs some smoothing).
- Need to write out sensitivity and distributed risk sections, model equations, and simulations. Need to add noise calculation to cooperativity. Then we can go back and trim down to ~5 journal pages. (based on refereed paper ~13 pages single spaced, ~25 equations).