10:30 A – 10:00 P, 11:00 P – 12:15 A
Probe extraction
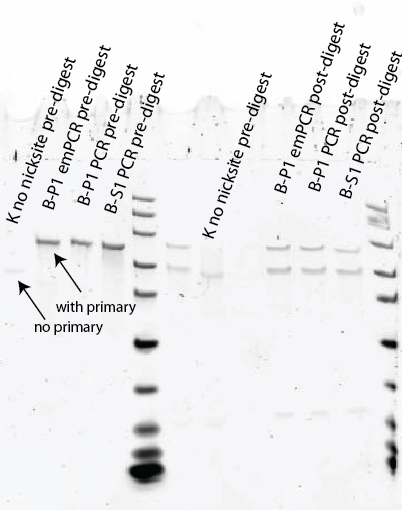
- Previous test gel, suggesting some samples at least have probe:
- Run denaturing gel at 100 uL scale 95 minutes. Cut out smallest band over 50 bp.
- sample loading order:
- ladder 5 uL ladder in 25 uL TBE + 30 uL 2x loading buffer, split into 2 30 uL loading lanes.
- B and K 30 uL sample + 30 uL freshly de-ionized foramimde loading buffer with 5% .5M EDTA.
- others are all 60 uL + 60 uL loading buffer
- back side: B, Y, K, G, Ladder
- front side, when flipped around: U, AA, AB, Ladder
Probe making troubleshooting
- column cleanup of emPCR and regPCR
- save first flow-through in case we need more product
- elute everything in 20 uL ddH2O
- 15 uL digestion reactions:
- 15 x (1.5 uL buffer, 1 uL enzyme, + 7 uL ddH2O)
- split 9.5 uL per tube x 13 tubes
-
- 5.5 uL elute DNA into each
- ND
- B-em PCR: 39
- y em-PCR 23
- ubx em-PCR 13
- B-PCR: 47
- Y-PCR 34
- U PCR 84
- K1: 42
- k2: 33
- K3: 1
- B-S1: 57
- Y-S1: 44
- K-S2: 50
- G-S1: 8 (maybe all 230)
- all ND have high 230 readings
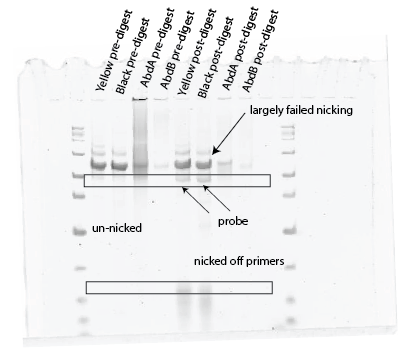
Gel, finally, good digest:
STORM
- Finish O/N STORM run of wt Ph-647, copy data to Monet (Alistair10)
- slide last night was actually wt Ph-FLAG m-647 + rb wt Ph-750 (no wonder it looked like previous wt Ph-FLAG)
- setup new O/N imaging of real wt Ph-647
Embryo labeling
- stain cryo-section embryo with m-anti-en-cy3 + rb anti-H3K27me3-A647
- Blocking
- staining secondaries, 11:45 P – O/N
- image with beads
- restain