10:10 A – 5:30 P
RNA labeling of cultured cells
- incubate cells in 55C RNA hybe buffer, 30 min
- transfer to RT 2x SSC, wash 10 min
- transfer to PBT wash 10 min+ 2x
- block 1 hour
- stain with shp anti-dig, 1 hr
- rinse
- block
- stain with 555 anti-shp
Cell culture
- Dilute cells 1:3 in new media, passage cells into new flask
- Move to 28C to grow until Thursday.
Probe making
- test gel.
- lanes not at all as expected
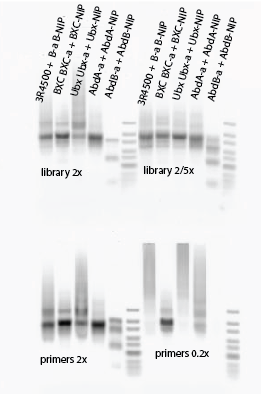
- new test PCR: Try different library concentrations and primer concentrations.
- library 2x (1 uL in 50)
- library 2/5x (dilute 1:5, then add 1 in 50 uL)
- library 1x + primer 0.2x
- library 1x + primers 2x
- Gel order: 3R, BXC, Ubx-a, AbdA-a, AbdB-a
- gel extract lower T7 bands (gel was taken by another?)
STORM
- finish imaging y-locus
- start imaging black locus. Spots much more visible and much longer lived when viewed in imaging buffer with Glox. (should try this with my new POSS).
- STORM switching of k locus pretty good, recording 85K frames.
- setup run. Recorded 3 positions, then aborted to test pyranose.
Imaging
- Make fresh make pyranose oxidase for pH stable switching: 6 mg in 100uL im buffer + 100uL catalase for 100x stock. Labeled as P.O.S.S. (Pyranose Oxygen Scavenging System)
- test POSS vs glucose on DNA. 1x POS = substantially less switching than 1x GLOX. 1/10 x POS is even worse. 10x POS is comparible. 10x Similar mass ratio to glucose oxidase, which I normally do at 18 mg per 100 uL.
