Thursday 07/25/13
STORM
- break down O/N run of R2bxcD
STORM analysis
- run all bxc data dot-fitting (que up on Tuck in small batches, Cajal not talking)
- run all bxc mosaic file conversions (que up on Cajal in linear batches)
Embryo staining
- incubating 2 coverslips of Cy3B-labeled En embryos in 488-WGA 1:600, 15 min
- rinse out WGA, 20 min
- incubate in poly-lysine
- wick off poly-lysine, quick rinse with PBS
- add PFA post fix 15 min
- Rinse in PBS 2x
- Scan complete coverglass to ID all embryos (4x, 488 channel)
- Scan all embryos for En-expressing cells. Scans started ~12pm.
- Scan finished 9pm
- using spacers from Shu, cut out to fit 22x22mm slides. Hopefully this + the crosslinked poly-lysine help keep the samples on the coverglass!
- accidently skipped prehybe, incubated in en-directly labeled 2 + unlabeled P1+S1-long+T1.
- removed coverglass and hybe solution, moved coverglass to prehybe
- remixed correct mix 40 uL hybe + 8 uL en-unlabeled-primary-r2 + 3 uL en-labeled primary-r2 (I know this works, this is the first batch of unlabeled primary r2 to be tested), + .5 uL of diluted 405 (previously way too concentrated, diluted stock down by half, adding another 200 ul), and 3 uL of secondary 1 with tertiary docking site.
- Prehybe 30 min.
- built new chambers. Much easier to use the whole thing than to try to work from a cut edge — gives more space to start peeling off the top as well. scraped away two sides on longside of coverglass, (we just want a spacer, not a seal. Will still use very thin layer of rubber cement to seal. This scraped off edge should allow better access to the tweezers to gently remove the coverglass after incubation.)
- add probes to slide. Wick off excess prehybe. Carefully invert slide ontop of chamber. Seal with rubber cement.
Chromatin paints library
- Redoing primer library to get 3′ G or C instead of 2 of last 5 GC.
- Also insist 3 or fewer of last 5 are GC (reduce mispriming events)
- BLAST P1 S1 and T1 sequences against primer library.
- HoxD library assembly
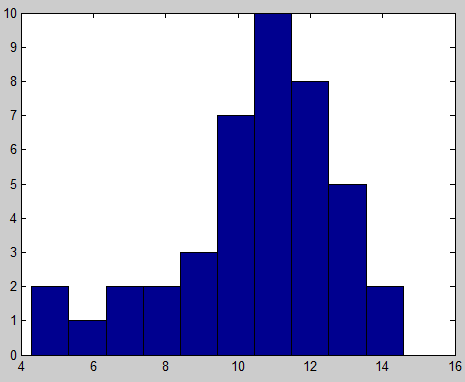
- distribution of number of probes per kb for all sub-libraries in library:
To do
- finish design of Lib 2
- ORDER lib2!
- Flip Fly stocks
- AbdA/AbdB/Ubx probe making
- review
This entry was posted in
Summaries and tagged
coding,
embryo labeling,
STORM. Bookmark the
permalink.

