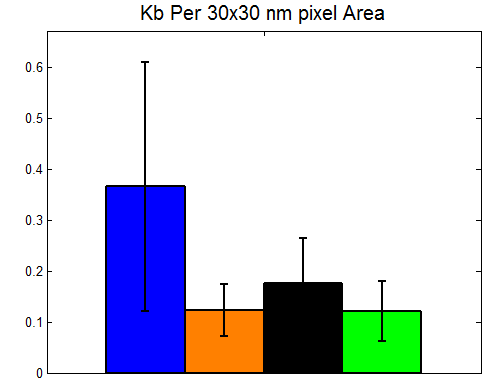
9:30 A – 7:30 PM, 8:30 P – 11:30 P
(volleyball 9:40 A – 11:30 A)
transcription modeling
- irreversible stacked cycles
cycle: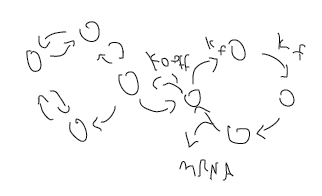
-
Once the system starts the mRNA production loop, there is no going back, it’s going to make an mRNA.
- probability of choosing this loop is p = kf/(koff+kf).
- distribution of the # of cycles (burst size) is Bernoulli(p).
- In this case, variance = p(1-p), CV^2 = (1-p)/p, goes to zero for p-> 1 –> kf>>koff.
- I don’t think this is right, the # of cycles can easily have an average greater than 1 (i.e. greater than p).
- probability of (exactly) n cycles is p^n(1-p) = (kf/(koff+kf))^n*(koff/(koff+kon))
Issues
- this is still the instantaneous burst assumption, that the kinetics of the this bursting process don’t affect mRNA distribution
- this guarenteed mRNA case is only true with no back reaction out of the first state.
Review writing
- read edited and aggressively trimmed review sections Introduction through beginning of transcription cycle modeling.
color chromatin data analysis
- For comparing distributions: http://www.mathworks.com/help/stats/kstest2.html