9:15a – 1:00a
Goals
- Slides for meeting with Levine
- Chromatin STORM analysis
- Analyze D11 region
- cells should arrive today — start in culture
- Library 3 design
Library 3
- observation: short green regions (<50 kb, basically everything not near a centromere/telomere) tend to be genic regions with extremely long exons and very short introns.
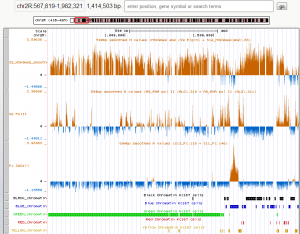
- Lam enriched regions 10-350kb in Kc167 cells
- chr2L:1994969-2008366 length: 13397
- chr2L:18991061-19001768 length: 10707
- chr2L:21683064-21738894 length: 55830
- chr2R:8090663-8144402 length: 53739
- chr2R:19706195-19726934 length: 20739
- chr2R:20581435-20630057 length: 48622
- chr3L:1250391-1295624 length: 45233
- chr3L:3102719-3114680 length: 11961
- chr3L:7154799-7231065 length: 76266
- chr3L:18903046-18983014 length: 79968
- chr3R:14754844-14804163 length: 49319
- chr3R:16852359-16879138 length: 26779
- chr3R:26653080-26707108 length: 54028
- chrX:840070-920095 length: 80025
- chrX:1381731-1406793 length: 25062
- chrX:6269580-6417829 length: 148249
- chrX:10223736-10238490 length: 14754
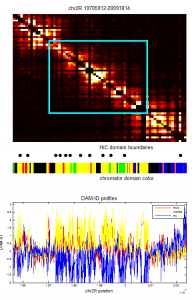
- Green regions not hard to find good isolated blocks of decent length (100 kb scale). Some pericentric regions are interspersed with yellow gaps, though whether these are really breaks or heterochromatic transcribed genes is not clear to me. Plenty of these regions are still H3K9me2 dense, unlike yellow regions elsewhere. (see image below).
- see Lib3PrepTable
- see chromatin scripts from today
- New code to plot Kc data for an indicated region and flanking spacer —
PlotKcData(locus,'spacer',spacer)
Genomics
- Gencode: Download GTF http://www.gencodegenes.org/releases/18.html
- ucsc/Encode, Experiment Matrix, A549 cells, RNA-seq. Look for Alignments, whole cell, pA+. Download BAM files for both replicates. (~12 Gb each).
Odyssey computing
- Directions for getting X-window window display forwarding working for odyssey: https://rc.fas.harvard.edu/faq/using-windowsmac-and-x11-forwarding/
- New Odyssey directory
\\rcstore.rc.fas.harvard.edu\homes\home05\boettiger
Using cufflinks on Odyssey
module load bio/cufflinks-0.9.3- Directions from Odyssey RC-computing
- 3 hours, still failed to get to run
Alternative approach
- Attempting to upload to Galaxy
- Penn State Galaxy uploads the file but freezes further operation because history size is too large (~12 Gb).
- Attempting to upload to Tubigen Galaxy server.
Ph model
- still runs slow. Still trying to get good range of plots for PhM concentration effects
Project 2
- working on write up for XZ.
Cell culture
- new cells arrived today
- morphology looks much better
- starting density not so bad. need to change media tomorrow to further remove DMSO. Hopefully these grow well from here out.
- Plan going forward — keep two separate stocks with two separate stocks of media.
- Freeze down a large batch of these cells a within two weeks from today.
Chromatin library deep sequence analysis
- Jeff wants to see binding motifs for off target sequences.
- looping over lists is slow
- this is going worse than expected… (1 am).