9:40a – 11:30p
Goals
- show Hao new project 2 results
- RNase-Away / bleach treat everything on my bench
- fix and stain new RNA cells. Allow 1 hour prehybe.
Project 2
- See notes
Chromatin colors, double stain prep
Probe organization
- reorganized both boxes of completed probes.
- Should start a new database in google drive to track completed probes, primer used, etc.
Region selection
- List of regions to make for double multi-color staining:
- G2, G3,
- G5, G6
- G1, G2
- F11, G1
- F12, G1
- D12, F11, F12, G1, G2, G3, G5, G6, + neg control
- Setting up PCR. Spec primers:
- at 20 bp (common), 10 uM is ~66 ng/uL. 40 bp is ~132
- dilute D12p is good, F12p is good, F11p is bit low, G01p 1:1.5 (add 50uL ddH2O to 100 primer), G02p add 0.8, G03p add 1.1, G05 add 1.4, G06p add 1.6;
- set up PCR to run overnight
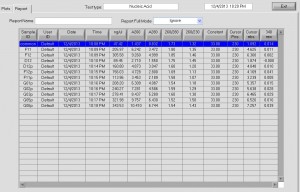
STORM analysis
- analyzed G1 and G9 data (80 kb blue ANT-C piece and 75 kb freestanding blue).
- G1 image quality degenerates over time (especially conventional images).
- suspect bleaching by TIRF beam. Discuss arraying image direction with Bogdan.
STORM
- E4 and E5 may not have worked. But then I was having issues with STORM4
- F12 (Taf) definitely worked, very clean dots.
- Imaging F12 overnight.
Cell staining
- Fix new cells
- prep cells for hybes
- prehybe 4 hrs at 47C in prehybe mix
- attempt RNA stains of D12, E08 and F06 (E10 basically empty). 3 uL D12, 1.5 uL of the others. 1 uL L2S1 (a647) in 20 uL hybe dilution mix.
- incubating at 47C ~10pm.
Chromatin Colors Data Analysis
Bead Processing:
- Running z-calibration for 647 beads for G09 data from STORM4
- Running chromatic calibration for G09 data from STORM4
- Analyze chromatic beads for G1
For future: Troubleshooting difficult probesets
- Prep for sequencing:
- good: G2, G3, D12,
- oddly difficult: G4, F3, F4, F5
- Wrote to Jeff to inquire about new optimized seq prep protocols.