9:50a – 12:00a
Goals
- label RT PCR
- set new RTs
- cast gels
- matlab-storm update and STORMfinder + STORMrender migration + demo
Probe making
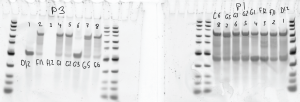
RT
RT round 3
- Run RT with ~5 uL RNA and ~1.2 uL primer 1 and primer 3
- cast new denaturing gels
- Test RT reactions on denaturing analytic gels.
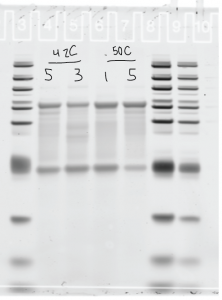
Probe clean up
- Combine previous PCRs: 7-20, 8-20, 7-40, 8-40, 1-pure-50, 2-pure-50
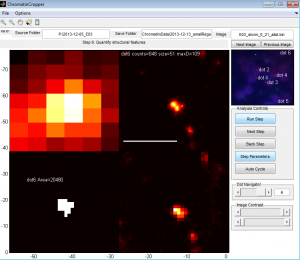
Chromatin Spot analysis
Code: matlab-storm
- moved STORMfinder and all its dependent scripts for out of Beta.
- moved STORMrender and all its dependent scripts out of Beta.
- synchronized master.
- switched back to alistair development branch for local work.
Ph Project
- need to adjust model figure:
- Put simulations of each model and the raw data plotted on the same axes.
- remove current panel A (that can be for the supplement)
- move panels D and E (polymer clustering models) to the supplement as well
- need to write caption for the modeling section.
- need to write main text / results section discussing modeling
- consider intronic FISH (or total RNA FISH?) for lowly expressed transcripts, see if some cells break through at high levels
Project 2
- Fixing issues with BLAST
- concern that low expressed isoforms of highly expressed genes may shelter off target sequences not tested in BLAST.
- Could build a BLAST library just concatenating all exons. We could give this names that match the genes.
- GAPDH has only 4 valid probes under current stringency conditions. Try weaker conditions