11:50p – 12:45a,
2:00a – 3:20a (remotely)
Chromatin Project
Data analysis
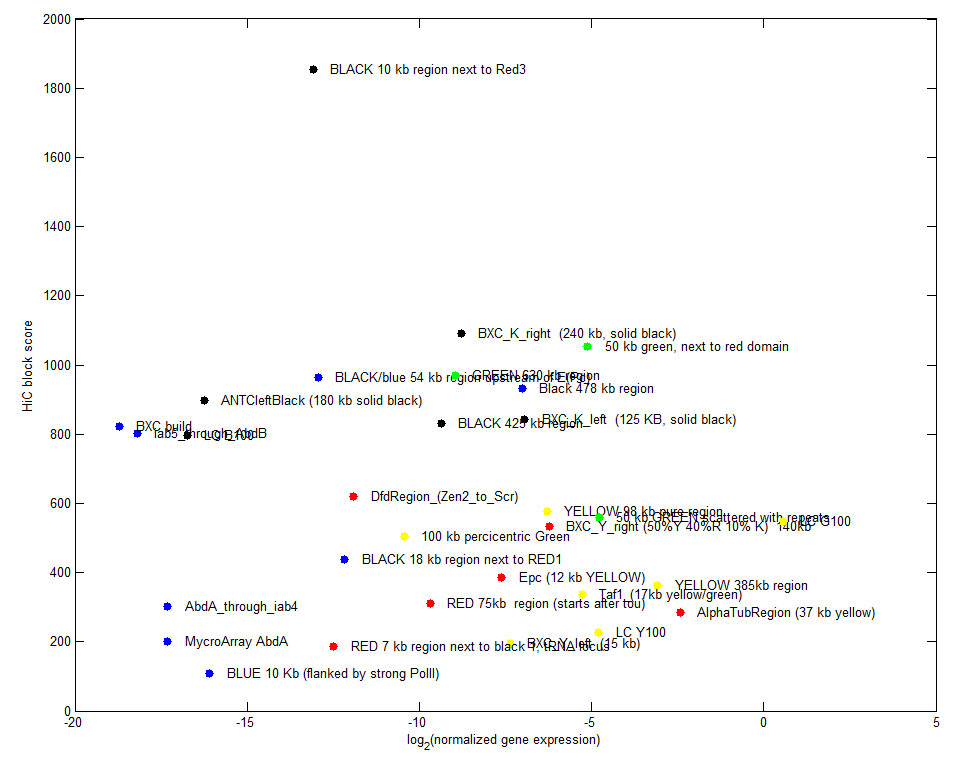
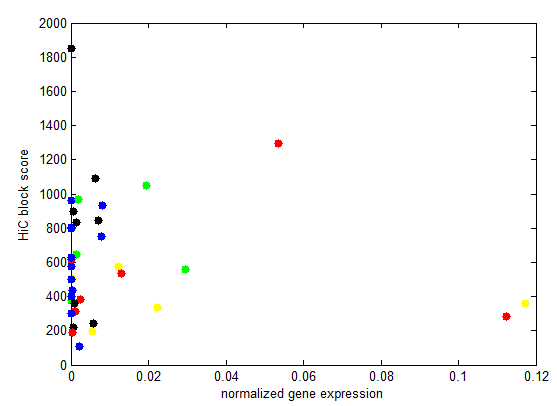
Goal: Sort data by expression levels.
Getting FPKM expression values:
Downloaded data from NCBI GEO database from the 25 Drosophila Cell lines paper.
Downloading gene name conversion tables from FlyBase
ftp://ftp.flybase.net/releases/FB2014_01/precomputed_files/genes/
This file is good: fbgn_annotation_ID_fb_2014_01.tsv. My earlier gene list has issues. No Antp.
CGs aren’t fixed length. Antp is CG1028, but lots of other genes are CG1028#. Uses strcmp.
Lots of new plotting stuff for libraries
Here’s a few examples, working out plotting genes and visualizing gene expression levels.
Some issues that need to be fixed still:
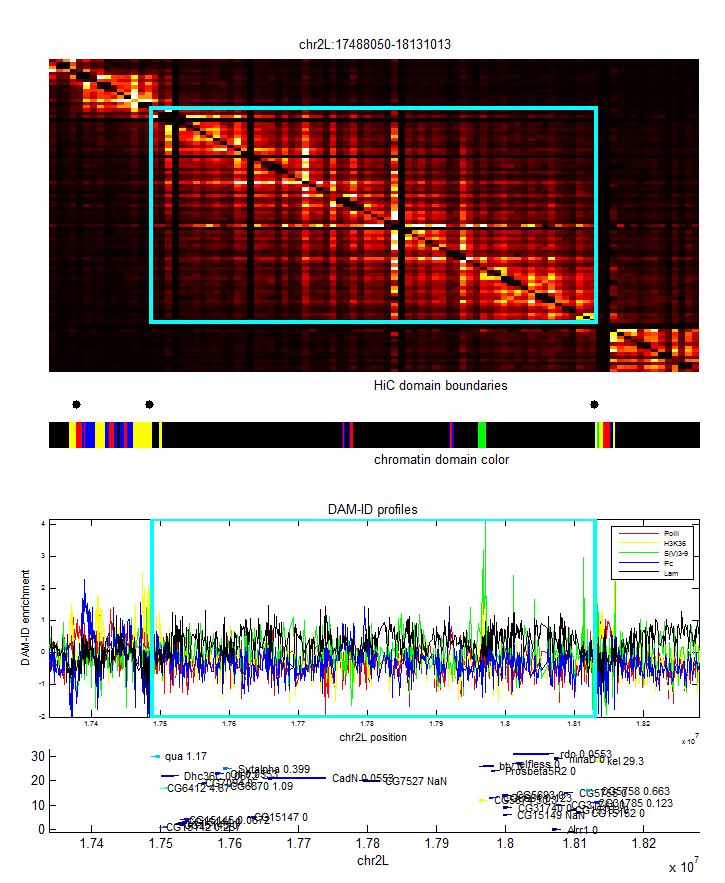
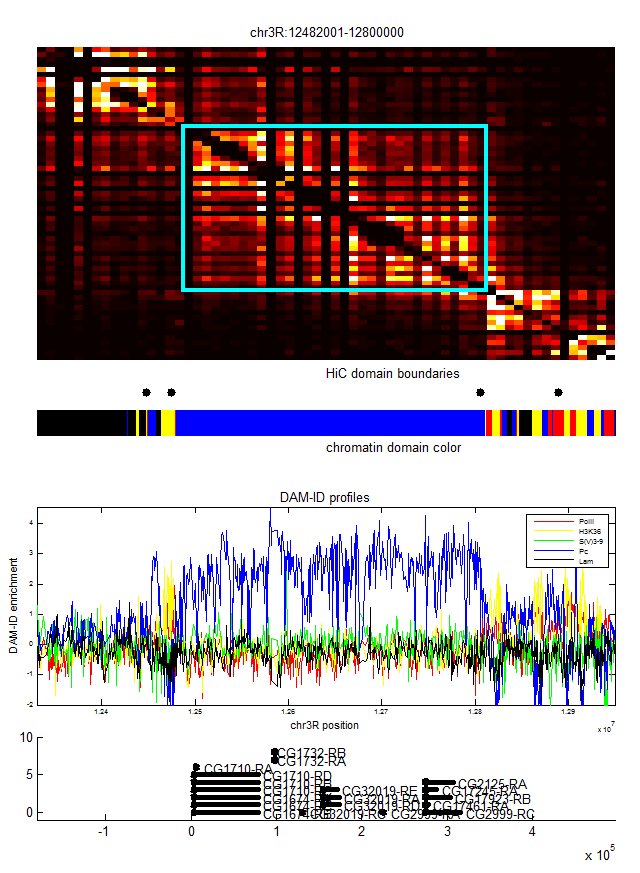
- match area names to table. This should be easy, original table is alpha-numeric. Only problem is the conjugate domains and the stuff from the previous MycroArray and LC libraries…
- GetLocusColor is buggy. Really just need to take the difference between the locus Start and locus End and the nearest boundary and record the chromatin type there. Not good enough to bounce out to the nearest interval boundary, this is less accurate and can severely distort things in cases.
STORM
- made fresh 1M MEA
- 2 uL MEA + 5 uL COT much much brighter A750 switching than previously. Need to repeat previous images with this stuff. Might need to always make MEA fresh (takes maybe 150 s).
- Bead concentration getting a bit low, time for new 1:1000 beads.
- stage movement is still cr*p. Need to manually pick regions until I can find time to get the USB interface to the nano Z-positioner working.
- sample imaging around 8p
- by the time the sample is arrayed and all the conventional images are taken, the buffer seems to have given out?
- 10:30p, buffer basically depleted — 750 switching is completely crp now.
- Maybe not — 2nd position looks better, maybe forgot to delete the calibration position from the position lists… beads were pretty dead there too…
- okay, switching alright still (12:35a). I’m going let this run overnight and hopefully the data’s okay. At least we have on-the-fly compression of the bead data now!