9:30a – 4:00p
Computer Management
- attempting to give users rights to share their hard-drives on the network without having full Admin capabilities to do anything they want.
- Tried adding my ‘User’ account to these groups
- “Performance Monitor Users” — can access performance counter data locally and remotely.
- This doesn’t actually let me launch task manager without Admin rights. Not too surprising, since I can kill tasks from task manager.
- Can’t even access the windows program “Performance Monitor” without Admin rights.
- Can’t launch the “Resource Monitor” without admin rights either
- “Access Control Assistance Operators” — windows / google seem to provide no information on what this really does.
- “Performance Monitor Users” — can access performance counter data locally and remotely.
- Can now open preferences, still get asked for admin password to try to change advance sharing file permissions
Chromatin Project
Data Processing
- SplitDax finished (hopefully for the last time) on E02 data
- running bead averaging / compression on E02 data
- DaoSTORM finished on E03 data
troubleshooting hybes
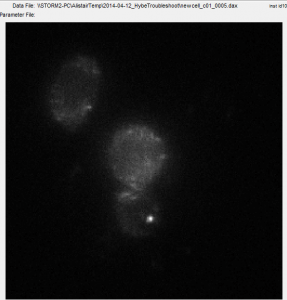
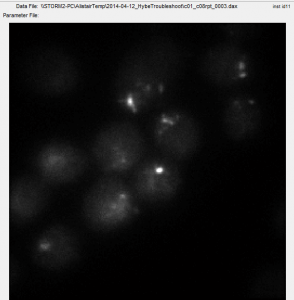
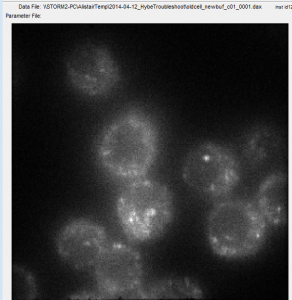
- have staining now of C01 in both new cells and old cells
- looks better in new cells
- neither is comporable to earlier stains or restained c8 cells.
- some cells have strong/decent stains and some cells lack staining.
- pretty sure there’s higher background, different quality. May relate to lack of RNase treatment in new cell round. —
More notes
- should have checked — C01 is 240 kb Y. We probably do have mRNAs in cytoplasm.
- don’t have spots in every cell, when we do have spots the cytoplasm is dimmer. maybe I’m just staining RNA. Can treat these cells and see if we lose stain?
- previous C01-C08 batch of cells were first imaged 1 day after I made new RNase solution, so I don’t have it for sure in my notes but I’m pretty confident these guys didn’t skip the RNase step. So current stains are consistent with not denaturing the DNA, except that a side by side re-treatment of C08 cells does denature for strong incorporation. So something is still different in the cell prep that affects the denaturation, rather than the denaturation itself.
- maybe RNA also dilutes out concentration of local probe (I kinda doubt it, probe concentration is pretty high since whole hybe soln is visibly blue).
- maybe somehow cells are over-fixed and won’t denature? I think I’m very consistent though on fixative and fixation times…
- I guess I often let them sit longer in the glycerol than the 30 min I have been doing — I sort treated that as a minimum to get properly equillibrated rather than an exact time step.
To try
- RNase treat current stained C01 new cells, see if foci are lost. Treatment started 1:20 pm.
- RNase treating new cells prepped with new buffers. Will then do extended pre-hybe.
- 95C denature of RNase treated cells prepped yesterday and restain with C01.