10:00 am – 12:45 am
More simulation troubleshooting
Experimenting with weaker attractive forces
energy = ("index*epsilon * .000000001 * (step(r-rm)*(r-rm)^4 * (1- step(r-2*rm))+ (rm)^4*step(r-2*rm));"
"index=index1*index2")
10 fold stronger bonding: “sticky8” simulation
energy = ("index*epsilon * .00000001 * (step(r-rm)*(r-rm)^4 * (1- step(r-2*rm))+ (rm)^4*step(r-2*rm));"
"index=index1*index2")
not very substantial change in structure
further 10 fold reduction
- sticky 9
energy = (“indexepsilon * .0000001 * (step(r-rm)(r-rm)^4 * (1- step(r-2rm))+ (rm)^4step(r-2rm));”
“index=index1index2″)
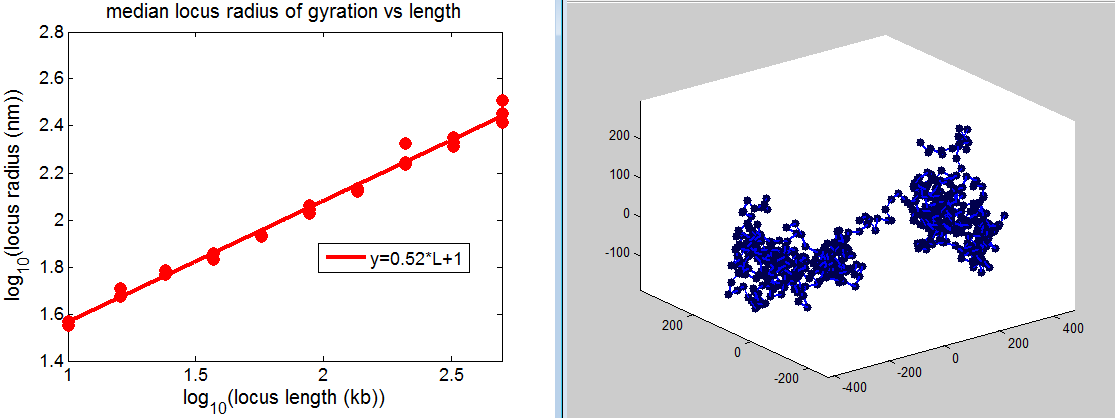
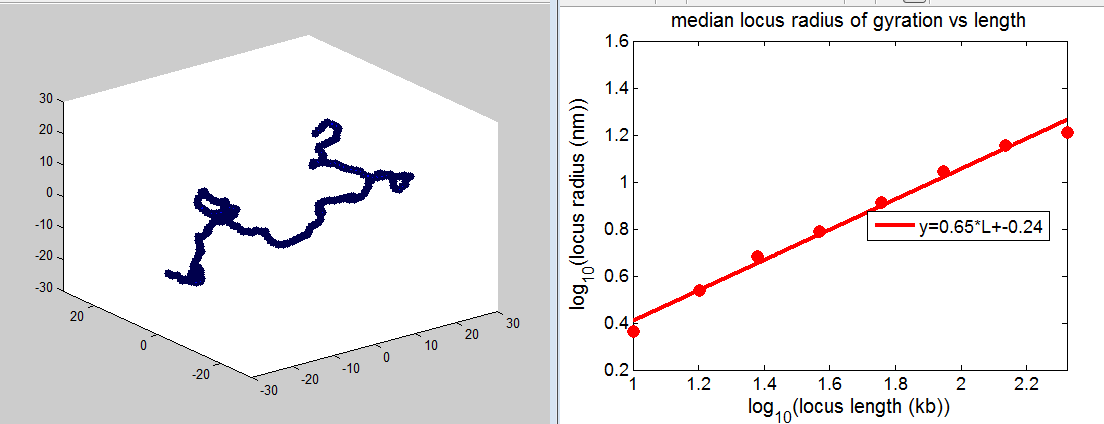
Despite the fact the scaling is only slightly below the SAW polymer, the system is not fully self avoiding.
The closest two monomers at any point throughout the simulation gets closer and closer as N increases, rapidly falling below the 10 nm exclusion.
redoing simulations with no attractive forces
- let’s check the self avoiding condition again.
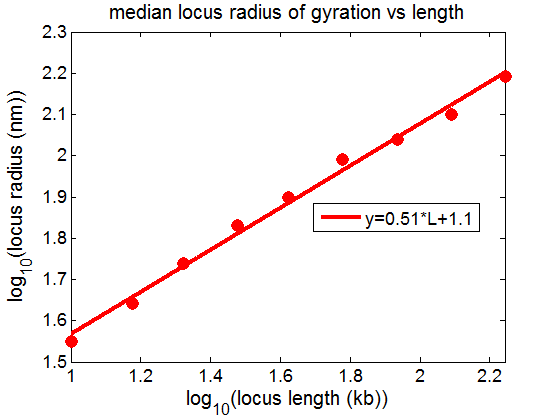
- hmmm… This actually looks very similar.
- and I thought this scaling came out to .60 last night but doesn’t seem to.
More issues
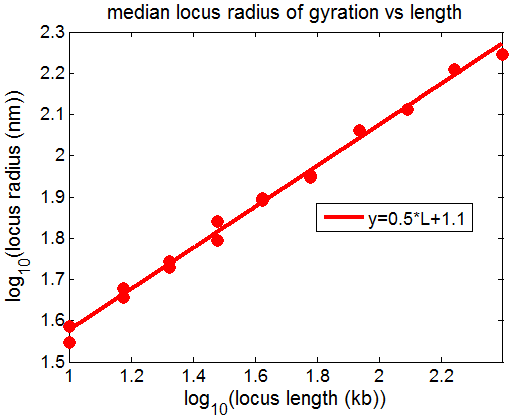
Now I can’t repeat Sticky6 (~flat scaling), final simulation to complete last night, in which I added a weakened attractive force back in after believing I had gotten the random walk polymer to scale properly at 0.6.
Mirny lab code
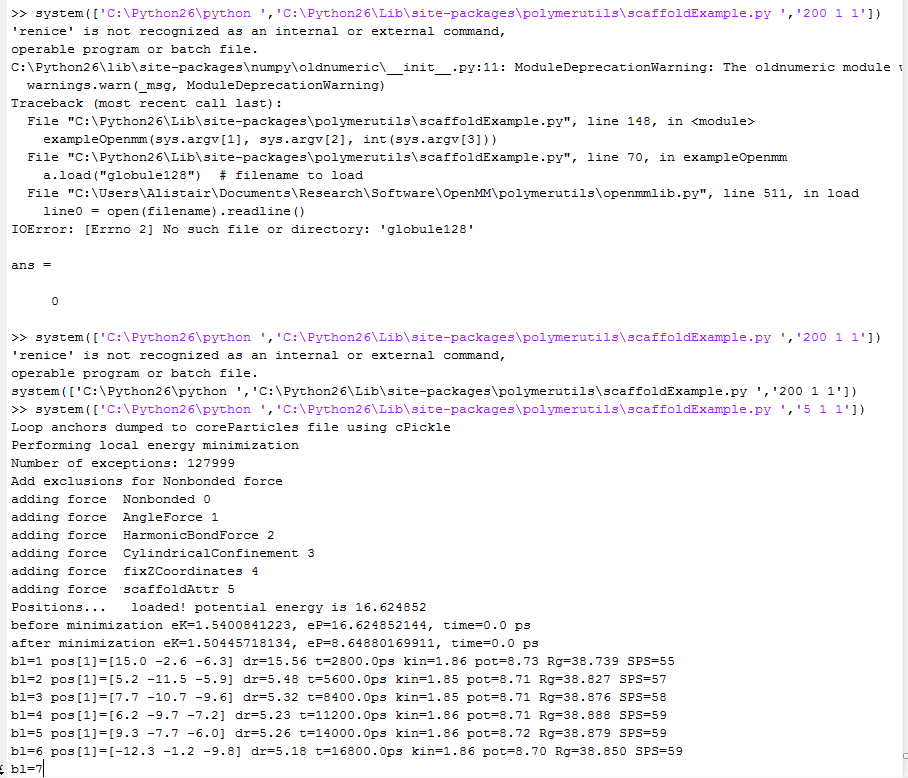
- Bashed away a bit at scaffoldExample to get it to run:
- This is very slow. But globule 128 is 128,000 monomers, so that’s probably why.
Trying to shorten simulation time by reducing data size. Wrote script to load and truncate
Finally got this working
- wrote a matlab wrapper to initialize the simulations using my RandomWalkPolymer function and to change key parameters like number of nodes and simulation time (see GitScripts)
- simulations seem to be behaving well:
Questions on polymerutils
- what are the inputs
exampleOpenmm(sys.argv[1], sys.argv[2], int(sys.argv[3]))the example has 200 1 1 but doesn’t explain what these numbers mean. - Can someone explain the shape of the Grossberg and LG ‘repulsive’ potentials? Why is there a minimum near the contact radius that goes up and saturates afterwards.