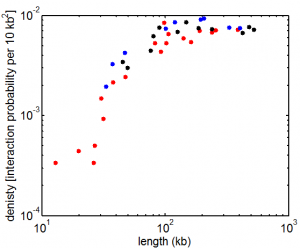
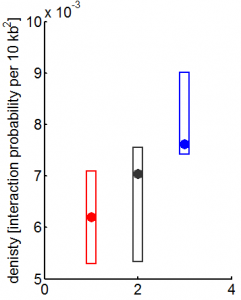
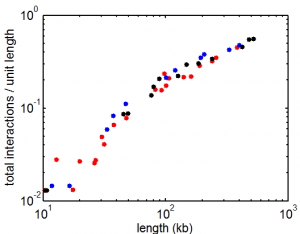
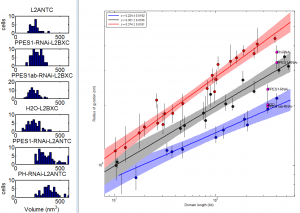
9:00 am – 8:00 pm
Goals
- prep new RNAi treated cells for staining
- Plate, fix and lyse current RNAi treated samples
- RNA isolation prep
- cDNA production
- qPCR reactions (?) [tomorrow]
- Set up synthesis reactions for new RNAi
RNAi experiments
new dsRNA synthesis
- ran PCR cleanup on new samples of Pc-v2, Psc-v1, Esc, SCM (2 PCR tubes each / 100 uL total reaction)
- set up T7 reactions:
- 20 uL eluted DNA-template
- 20 uL dNTP buffer mix
- 4 uL T7
- 4 uL RNasin
- running for 16 hrs at 37 C
RNA isolation
- following Qiagen RNAeasy kit
- the second round elution does make a difference (1.25 – 2x more yield)
- a third round elution does not help. temperature of water for elution does not matter (tried 4C and 37C)
sample order
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 1
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 2
- Pc v1
- Pc v2
- Pc, Esc, Scm,
- Esc
- Suz12
- Scm
- SCE
- mock
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 0 (from last week)
* concentrations in the 300 to 800 ng/uL range
First strand cDNA synthesis
RNA dilutions (master 1 12x)
- (desired 10 ug of RNA) -> 20 uL RNA (have now ~200 ng/uL instead of 2500 ng/uL)
- 2 uL oligo-dT primer (24 uL)
- 2 uL dNTP mix (24 uL)
- heat to 65C for 5 min, return to ice.
per reaction (master 2, 12x)
- 4 uL 10x buffer (48 uL)
- 8 uL MgCl2 (96 uL)
- 4 uL DTT (48 uL)
- 2 uL RNase OUT (24 uL)
- 2 superscript3 (24 uL)
sample order
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 1
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 2
- Pc v1
- Pc v2
- Pc, Esc, Scm,
- Esc
- Suz12
- Scm
- SCE
- mock
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 0 (from last week)
final step
- add 2 uL RnaseH to each reaction
- I’ll set up the qPCR reactions tomorrow.
- samples in -20C in front part of 8-strip rack.
Cell prep for staining
- prepping all PPPES slides and wt controls for FISH
- most of these slides have almost unusably low cell denisty
- staining slide 1 PPPES (0) for Antp-P1 (2 uL + .8 uL S1) O/N.
Prep new cells
- RNAi from 4 days ago ready to be fixed.
- Plated 8 wells of new PPES (top for are PPPES rep-1, next for are biological replicate PPPES rep-2), + 2 WT/mock + 2 PES (Pc, Esc, Scm)
New dsRNA synthesis
- Template making
- PCR strip order:
- Ph-p, Ph-D, Ez-v1, Ez-v2, Esc-v2, Psc-v2, Psc-v3, SCE-v2
- running O/N
New RNAi knockdown
- 3 wells of 6-well plate with 30 ng Ph-P and Ph-D
- 3 wells of mock
- not 100% sure I added the serum free media for the serum shock… Will see when we check the qPCR I guess.
RNAi planning
- ph looks good, let’s push ahead with this as a single.
- discussed doing RNA-DNA doubles. RNA with dig in conventional, antibody stained and post-fixed prior to digestion.
- will try this soon — could use a P1-dig