10:15 am – 8:00 pm
RNAi experiments
Quantifying knockdown efficiencies by qPCR
cDNA
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 1
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 2
- Pc v1
- Pc v2
- Pc, Esc, Scm,
- Esc
- Suz12
- Scm
- SCE
- mock
- PPPES (Pc, Ph-p, ph-d, Esc, Scm) rep 0 (from last week)
genes to test
- Pc
- Ph-p
- Esc
- Scm
- Suz12
- SCE
- Abd-B
- Antp
- Ph-d
- Gapdh1
- Act5A
Experiment layout
- Note: don’t have primers for Su(Z)12
- see qPCR setup google doc
- 2.5 uL of primer mix
- 2 uL of cDNA
Results
- knockdown doesn’t seem to have worked so well this time.
- maybe some issues with the qPCR, will try to run a few lanes again.
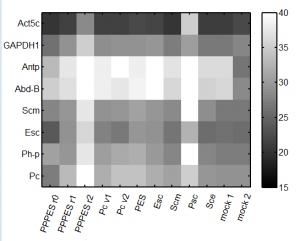
- Notes
- col 9 has Psc instead of Scm
- col 10 has Sce instead of Scm
- col 12 has ‘Psc, Sce, Ph-D and alpha-tub84a’ instead of ‘Abd-B, Antp, Gapdh1, and Act5c’ respectively
new dsRNA synthesis
- freeze T7 reactions of original PPES combo. Not sure we’re coming back to this.
STORM
- cells very sparse.
- note: the top filter wheel sometimes gets turned. It needs to be in position 1. take off the inspection port. if empty port 3 is visible in the inspection port, the correct port 1 is aligned. The IR focus lock beam bounces off this dichroic. No idea why people swap this out.
New stains
- prepped whole plate of cells
- should make initial density lower still — a lot of cells detach in sheets during the LN2 treatment.
- seems to me the PcG RNAi treated cells detach more than the wt / mock cells do.
- RNase treated just 2 coverslips. Rest still RNA-intact.
