9:15 am – 7:15 pm
PRE mapping
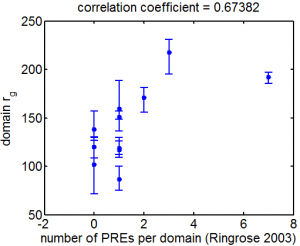
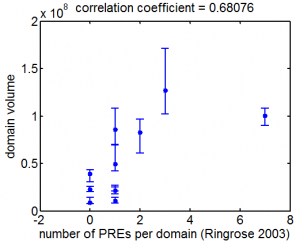
- Ringrose 2003 predicted PREs http://www.techfak.uni-bielefeld.de/~marc/pre/hit_list.html
- Ringrose 2003 is dm1. Need to lift over to dm3 before we can map.
- lift over, plotted correlation.
- serious computer crash (windows froze while unpacking .gz file), matlab history corrupted,
- not too strong a correlation with the motif derived ChIPseq data.
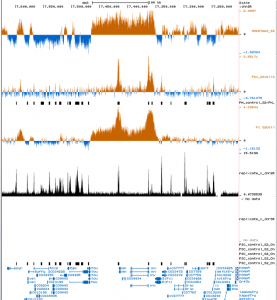
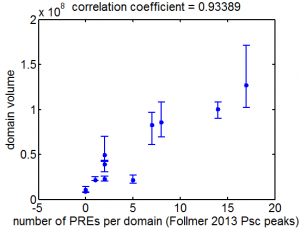
- using peaks from Follmer and Francis data
- these peaks look less PcG specific, the ones in the PcG’s match the ones I’d infere from modENCODE
- here’s the correlation:
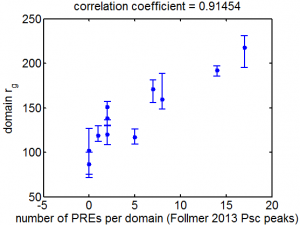
- should comment on pairing dependent silencing and (more importantly) multiple insertion silencing (see Kassis 2013 review).
Cell Prep
- poly-Lysine coated 24 coverslips in 12 well chambers
- plated Ph-p Ph-d RNAi KD cells onto coverglass 10 per plate + 2 WT/mock controls
- fixed cells, now in PBS in 4C
RNA isolation
- 2 1.5 mL eppendorfs each
- ran the Qiagen RNeasy extraction
- high concentrations this time (Ph ~1600 ng/uL and mock at 1400 ng/uL)
Maxima cDNA first strand synthesis
- protocol
- per reaction
- (2x the recommended mix for a 40 uL reaction)
- 3 uL 100 uM primer (= 300 pmol)
- 3 uL 10 mM dNTPs
- 10 uL RNA
- 14 uL ddH2O
- heat to 65C for 5 min
- then add per reaction
- 8 uL 5x RT buffer
- 2 uL enzyme
- 2 uL RNasin
- mix, run at 50C 1 hr, then heat denature.
- froze extra RNA at -80C
- cDNA reaction still running.
Ordering
- ordered finer forceps (I the cheap ones I bought are too stiff and I keep dropping my coverslips)
- ordered more coverslips
