9:20 am – 10:00 pm
Reading
- I’m looking for new insights to improve our genome wide prediction of PREs
- Schuttengruber et al Cell Reports 2014 on PRE evolution
- Zheng et al 2012 develop computational predictor
- this uses ChIP data to ‘validate’ predictions
- ChIP/damID data from Tolhuis et al (225 genes), Schwartz et al (176 genes), and Schuettengruber et al (215 genes) show only ~30 % agreement (38 genes)
- they remove ‘duplicate genes’ from list. NO!! I want to count multiple PREs per gene as multiple PREs. don’t remove the “duplicates”, they
- I want to see ChIP data used as input and PRE genetic tests used as validation.
- ROC curves for this predictor outperform previous model (jPREdictor) but are still not very far off the diagonal.
- back to Schuettengruber…
- Schuettengruber 2014 predict 379 conserved sites within PcG domains using cross-species ChIP-seq K27me3, K4me3 (for TSS), Pc and Ph.
- downloaded table
- NOTE: should plot with and without peaks corresponding to TSS’s for estimated PcG density.
- nice paper. weak, multi-component interactions specify PREs / PcG silencing, highly conserved through D. vir.
- also has higher res Hi-C than previous Sexton et al 2012, and focuses on PcG regions.
- claim PHO sites preferentially contact each-other uniquely in the context of PcG domains
- show predictive correlation and KD evidence that PRC1 recruits Pho (in Ph mutants). Specifically reduce Pho binding at its PcG sites but not its non PcG domain sites.
- mutants correlate Pho binding and Ph motif within PcG domains but wt does not (supporting cooperative recruitment model).
- outside of PcG context, Pho co-localizes with CP190 and BEAF32
Review
- finish refereeing paper and submit recommendations (due tomorrow)
Chromatin project
for analysis of deviants, comparison to other features
- getting D mel embryonic gtf file to run cufflinks
- SAM format specs
- SAM files need to be sorted first by SAMtools see thread.
- data is not presorted the way cufflinks needs, (should be alpha-numeric by chromosome).
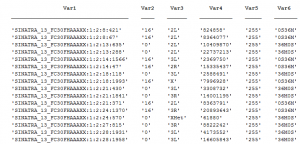
- sorting using this command:
sort -k 3,3 -k 4,4n hits.sam > hits.sam.sorted - (From BioSTAR: The code just means to sort on column 3, then by column 4(numerically) of the hits.sam file and print to hits.sam.sorted)
e.g.
sort -k 3,3 -k 4,4n /n/home05/boettiger/Genomics/Data/GSE18040_Dm_KC167.sam > /n/home05/boettiger/Genomics/Data/Dm_Kc167.sam.sorted 2> /n/home05/boettiger/Genomics/Data/errorsSort_KC167.txt - best to test these things on small data sets
- wrote matlab command to build smaller dataset (ParseSAMdata_150727.m)
- sort command works as expected on this.
- this runs properly through cufflinks (tested small version)
- sorting the whole 3Gb data set on Odyssey with this command is very slow…
- sorting finished, running cufflinks still failed. Upset about ordering of chr M and chr U in the SAM file (neither of which I need!!)
- Moreover this file IS sorted correctly, U is after M (and before X and Y) so shut up and keep analyzing!
'current' 'hit' 'is' 'at' 'U:3652,' 'last' 'one' 'was' 'at'
'M:18987'
- samtools sort doesn’t work on sam files, only bam files (so much for “sam”tools).
- file REFUSES to convert to BAM becuase there is no @SQ lines in the header.
- okay, so let’s sort by hand and remove the ‘M’s and ‘U’s using matlab
- matlab textscan reads this into inefficient cell arrays, which are now using ~60 Gb (yes gb) just to textscan in a ~4 Gb text file.
- Bogdan is going to fix this in Python
- after some more frustration, data ran correctly.
RNAi
qPCR
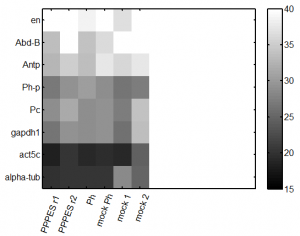
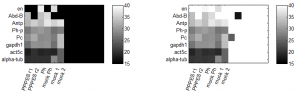
- setting up qPCR of last weeks PPPES (1 and 2) and Ph KD samples, along with corresponding mocks.
- assay for 3 cntrl genes, 3 PcG targets, + Pc and Ph-p.
- column order (cDNA): PPPES 1, PPPES2, Ph-Kd, Ph-Kd-mock, PPPES-mock, prior-mock
- row order (primers): alpha-tub, act, gapdh, Pc, Ph-p, Antp, Abd-B, en
- flipped primer labels. oops. fortunately I sorted by expression so it’s easy to spot.
Embryo staining
- check samples on confocal
- no staining at all.
- maybe 37 C is necessary.
- previous results look vaguely more encouraging
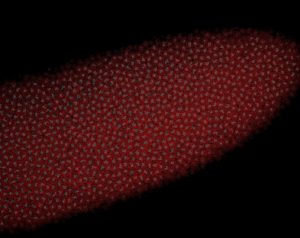
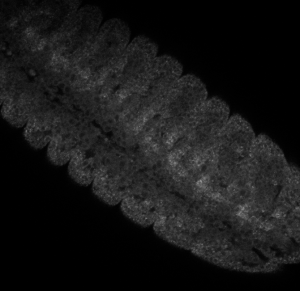

Issues with protocol
- Temperature not mentioned. I assume this means RT but I find that a bit strange for hybes
- probe sequences, probe length, and probe number not mentioned
Other
- gave 8 uL of 40ng/uL YW gDNA to AC.
