Purpose
- to start MERFISH experiments in Drosophila embryos
- Plan: use Biosearch probes to optimize RNA labeling using DNA probes
- currently using 20mers directly labeled. These aren’t working.
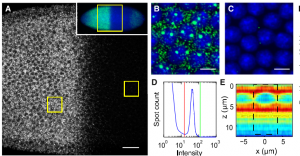
Images from Shawn Little’s paper (Gregor lab)
My RNA hb stains
zoom out on whole embryo, stained in green for hb
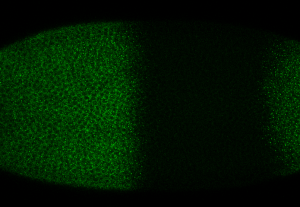
a different embryo with hb in red and an intronic reporter for a hb transgene (at a different insertion site on a different chromosome) in green
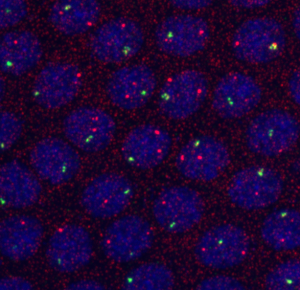
My DNA hb stains don’t work at all (following protocol from the Little et al Cell paper)
- Not clear why
- could repeat RNA stains again just to make sure everything is working alright. Not sure if my RNA-probes are still good after 6 years in the -20C but I can try.
- try different hybe conditions (tried RT hybe, which is what appears to be what Little et al use, tried 37C hybe).
- could try different hybe buffers
- I recall Raj reported not being able to get Drosophila embryos to work with his 2008 protocol