9:30 am – 7:30 pm, 9:00 pm – 11:45 pm
Chromatin Project RNAi
RT reactions
- protocol
- 5 uL gene-specific primer mix
- 10 uL total RNA, (diluted to 400 ng/uL)
- 5 uL RT buffer
- 1 uL dNTPs
- 1 uL Maxima
- 2 uL RNasin
master 10x
- 50 uL RT buffer
- 10 uL dNTPs
- 10 uL Maxima
- 20 uL RNasin
notes
- noticed precipitate in my RT buffer. opened a new buffer. thawing now.
- see same thing in new RT buffer. Didn’t notice before, but it’s pretty subtle
- Primer mix: alpha-Tub84, Act5C, GADPH1, Pc, Ph-p, Antp, Abd-B, en
- sample order:
- Ph 7-7 10 uL, –> 250 ng/uL
- mock 7-7 10 uL –> 250 ng/uL
- Ph 7-14 2.5 uL –> 210 ng/uL
- mock 7-14 2.5 uL –> 210 ng/uL
- Ph 8-8 10 uL –> 210 ng/uL
- mock 8-8 5 uL –> 450 ng/uL
- Pc 8-8 5 uL –> 250 ng/uL
- mock 6-29 2.5 uL –> 250 ng/uL
RT clean-up
- try alkaline hydrolysis followed by oligo column-clean-up.
- eluted in 20 uL. Spec’d all samples. conc’s above
- dilute 5 into 20 uL ddH2O. use 2 uL per sample to set up.
- try running with a no primer control
QPCR
Reaction set-up per well
- 2 uL diluted DNA ~50 ng/uL
- 1.25 uL EvaGreen
- 12.5 uL Phusion 2x master mix
- 2.5 uL primer combo (fwd/rev)
- 6.75 uL ddH2O
Master mix (80x)
- 100 uL EvaGreen
- 1000 uL Phusion 2x master mix
- 540 uL ddH2O
- 20.5 uL per well
Plate setup
- 8 DNA samples + 1 no template control
- 8 gene-primer combos for each
- 72 PCR reactions
Analysis
- failed!! essentially indistinguishable similar amplification in all wells. We should have clearly orders of magnitude separated samples
- probably accrued too much PCR product for these reactions in the pipettes etc.
Gel-troubleshooting
- complete nonsense:
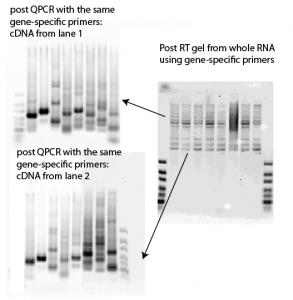
- stupid me. I mixed the foward and reverse primers already for the PCR. Can’t use both forward and reverse primers in the RT, this creates a mess.
To do still
- upload review!
- analyze SIM data
Other stuff done
- Psc null cells, finished freezing down stock
- send back shuttle form for EMBO -DONE
- checked staining on BXC cell cycle — not that great via turnkey. Should analyze this data further
