9:45 am – 11:30 pm
To do
- submit review
- book flights for EMBO (travel form / shuttle requests due on Friday)
Chromatin project
centromeric heterochromatin
- downloaded dm3 heterchormatic assemblies from UCSC here: http://hgdownload-test.cse.ucsc.edu/goldenPath/dm3/chromosomes/
- mapping previously imaged GREEN regions back to the chromosomes
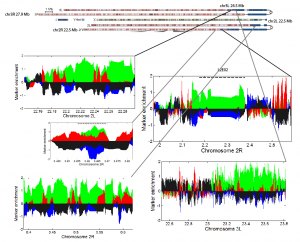
- download modENCODE chip data for heterchromatic regions
- downloading data from modENCODE: http://data.modencode.org/?Organism=D.%20melanogaster
- checking modENCODE data for agreement. Here is BXC for K79me3 (red), K27me3 (blue), K9me2 (green)
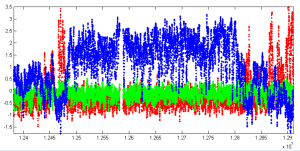
- compare to van Steensel data we’ve been using:
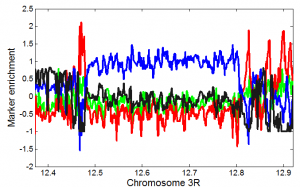
- looks pretty good, let’s go on with this.
- finished designing and assembling library. Will order tomorrow.
dual color labeling
- 750-P3-BXC + 647-P1-L2F6 no spots 🙁
- abbreviated wash. Try washing longer.
- try G5 G6 instead
STORM
- 750 laser desktop configuration file not running properly.
live imaging
- construct 1: mmaple3-MCP in H293 cells
- looks good! we see some telomeres
- construct 2: mmaple3-PCP in H293 cells
- also looks good, a bit more backgroundy
- construct 3: PCP-tagRFP in H293
- looks good. maybe best yet (though not STORMable)
- took some movies (called tagRFP) can estimate positional stability.
- construct 4: U2OS cells, gRNA co-transfect. TagRFP-PCP
- a lot of uber bright nuclei with nuceleolus stain
- not so good
