11:00 am – 7:45 pm
RNAi QPCR
- Oligo cleanup purified T7 DNA
- (probably could have done a standard DNA cleanup, I don’t need the supershort 30 bp stuff)
- ran plate of QPCR
- running out of primers.
- Should order more primers for Pc, Ph, Antp, en. (done)
- would be good to add some extra versions in as well.
- still need new primers for ph-d at least
- results look reasonable. Ph KD not at its highest
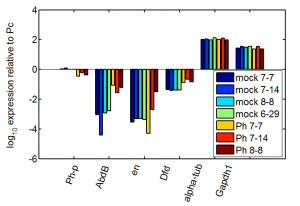
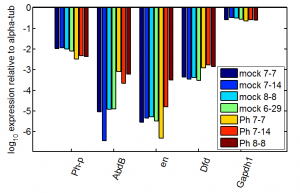
previous data
- this looks better with the raw DNA from yesterday, just I screwed up some of the primer labels
- maybe can repeat again when I get new primers in next week.
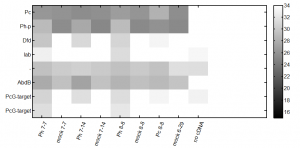
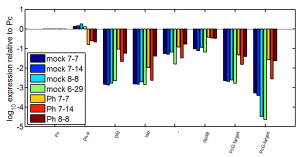
- Ph KD efficiency = 0.87 +/- 0.02 (3 biological replicates, mean +/- SEM)
new RNAi
- set up new RNAi for Ph + mock at large scale
- plus culture maintenance
- passage Kc cells in SFX
- passage Kc cells in Sniedner’s
- passage Psc-deletion cells
- all cells will be used for RNA extraction and sequencing
RNAi Ph quantify structure effects
- stain single-color control region for with new mock cells
- stain new ANTC-P3 + G6-P1
- stain new BXC-P3 + F6-P1
- these P1 probes are both good and should be decent controls for staining
- check staining on alternative BXC-P1 + F6-P3 this evening.
- this one also failed
- heat block is definetely mis-behaving.
- might have screwed up this hybe too — temp dropped 2 degrees C when I added the slides
Ph KD quantification: New immuno staining
- stained Ph-KD cells and mock cells with primary (1 hr RT)
- rinsed 30 min
- re-block (ready for rabbit secondaries).
