9:45 am
RNA library prep
setup
- protocol from NEB
- discuss plans with Jeff
- request training on BioAnalyzer and/or TapeStation instrument for library QC
- collecting kit components
RNA purification
- run out 1 uL on a 1% agarose gel in TAE
- treat samples with NEB DNase I (1 uL in 20 uL reaction, used RQ1 DNase buffer, don’t have an NEB one. validated the RQ1 is also a DNase I from Promega, guaranteed RNase free)
- Purify samples on AMPure RNA XP beads.
- performed all washes on column
- gel results: (clearly degraded). RNA is a month old (stored at -80C, but used for several RTPCR runs.
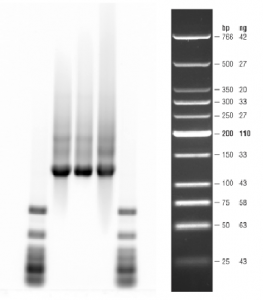
- probably do better to start 1 clean fresh run with newly isolated samples.
- we can run through the procedure today with these samples to get familiar with the protocol.
First strand synthesis
- ran without actinomyocin (don’t have any). Already did DNA digestion so hopefully this isn’t necessary
Second strand synthesis
- setup is easy
- run bead cleanup using Aline beads from Jeff in place of AMPure XP beads
More steps
- purify dsDNA (beads)
- end prep
- adapter ligation
- purify DNA (beads)
- this uses a lot of beads. Ordered more Aline beads from Aline biosciences
- PCR enrichment
- ran on QPCR machine with EvaGreen
- 14 cycles, started to saturate, pulled off
- testing sample by running out product on a gel
- Typhoon isn’t working — images are perfectly 100% blank. no noise, no contrast, no background of gel.
- still have 1 more bead clean up to do.
STORM analysis
- compute z-calibration from surface beads for STORM2 for Ella
- cancelled STORM imaging for tonight (insufficient time to setup a run before midnight, insufficient time to collect 2 color data if the staining works before 10am).
- signed up for STORM4 for Thursday and Friday for live imaging with Steven. STORM4 might not work — the 100x objective went missing.
Ph KD immuno
- finished staining
- tested samples on Turnkey. WT shows decent staining. Ph in KD not detectable, but need nuclear counterstain to validate cell region.
- maybe try imaging these on confocal tomorrow.
Editing
- Editing recommendations for YF application
- Editing Ph manuscript draft
- got new Ph manuscript draft, will try to add to current one.
