9:45 am – 8:00 pm,
remotely, 9:15 pm – 11:00 pm
RNAi validation
Finish library prep
- run final bead purification and elution
background prep
- contact Bauer core to confirm dates for tape analyzer
- reading background for KAPA_Library_Quantification
- stresses don’t freeze thaw repeatedly dilute DNA samples
- keep dilute DNA samples in a buffer, otherwise they degrade rapidly
- avoid multichannel pipettes and multiwell plates, but run lots of replicates (not convienent)
- reading protocol for KAPA Library Quantification kit
- got Kapa master mix from Jeff (has primers added)
setup
- 6 standards + NT control, 3 replicates (21)
- library dilutions (3 libraries)
- dilute each library 1:100. Lets do 2 library + 198 buffer. These are the working dilutions (200 uL).
- target dilutions: 1:800; 1:3,200; 1:128,000
- 10 uL working-dilution + 70 uL buffer (1:8 of a 1:100). total 80 uL
- 20 uL previous-dilution + 60 uL buffer (1:4 of 1:800). total 80 uL
- 20 uL previous-dilution + 60 uL buffer (1:4 of 1:3200). total 80 uL
- do this twice from the same working-dilution for each library (2 reps x 3 dils x 3 libs) = 18
- setup in 96 well plate (39 samples, 3×7 and 3x(3*2)).
- all dilutions still above the concentration range of the standards. Should repeat setup.
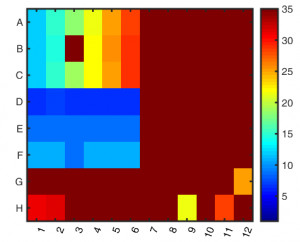
- some pipetting inaccuracies also detected.
Imaging
- staining Telomere-GFP samples with Draq5
- added Draq5 to Ph-KD and mock-KD cells stained previously with Ph-Cy3B
- quantify samples on confocal LSM700 (11am – 1pm)
- stained Ph-KD and mock-KD with Hoechst. Looks better than Draq5 on fixed cells.
Editing
- finish comments for Ye. Send back.
Coding: Quantify Fixation GUI
- adding alignment stats to GUI step 2
Data analysis
- analyzing internal data for BX-C L4E22to24
