9:30 am – 9:00 pm
Goals
- ChromatinCropper2 two color data for ANT-C G6
- Spot finding for two color BX-C F6 data
- ChromatinCropper2 two color data
- tape station of sequencing prepped library for Ph-KD and control
- qPCR of sequencing prepped library for Ph-KD and control
STORM
- stop STORM run, clean up STORM2 for Yari
- analyzing data on STORM2
- shouldn’t launch more than 30 jobs at once due to read/write limitations through the USB3.0 port on Morgan — makes interacting with the disk impossible.
Data Analysis
- troubleshooting issues with chromatic alignment
- fixed issues with chromatic alignment
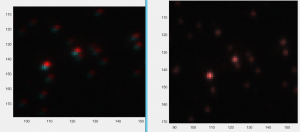
- Cropper2 not applying warps properly: previous warpfile was still hardcoded, not being passed
- fixed warp for alignment of conventional images.
- still need to make sure warp gets applied to STORM images.
Deep seq prep
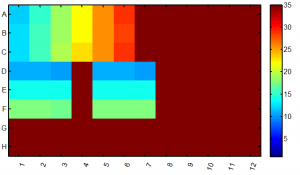
- rerunning Kappa quantification
- kappa standards 1:6 in triplicate (A1-6 through B1-6, C1-6) + No template controls A7-C7
- original dilution 1:100
- serial dilution (in triplicate)
- 1:100,000, 1:1,000,000, 1:10,000,000
- 10 in 990.
- just used median in fit. linFit(x) = p1*x + p2; p1 = -0.3005 (-0.3083, -0.2927), p2 = 4.989 (4.823, 5.156)
- looks good
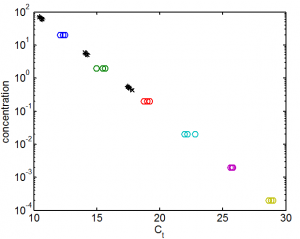
- 1 in 10,000 dilution is ~60 pM. so original is 600 nM, assuming average molecule size of 452. Otherwise correct by multipling by 452/aveSize.
Genome analysis
- figured out header structure and wrote missing header for modENCODE Kc167 SAM data.
- saved as dm3_SAM_header.txt on Morgan on
\\MorganData1\Chromatin\2015-08-25_KcENCODEdata
