11:00 A – 10:30 P
snail analysis
- Finish O/N confocal of MP06Hz
- Finish O/N confocal of MP08Hz sna[D]
- extract images from lsm,
- analyze nuclear density and record staging
- run dot-finder to count mRNA
- MP08Hz conclusions: 2 def. mutants in mid cc13 (counts are lower than wt cc13 starting levels). All 4 telophase embryos are wildtype (slightly higher than wildtype levels due to extra MP08 loci).
Fly work
- collect virgin
- sim[D] (keep in case need more crosses to test recombinants)
- Espl[D]
- yw; Sp/CyO,HbZ; Pr/TM3,HbZ (1 male only)
- Sp/CyOZ; TM2/TM6 (cross both ways to Esc[6]/CyO)
- Esc[6]/CyO (crossed to above)
- MTDs
- More crosses:
- mBT x MTD
- Psc shRNA 1 x MTD
- Psc shRNA 2 x MTD
- Pc shRNA 2 x MTD
- Rad21 shRNA x MTD
- Su(Z)12 shRNA x MTD
- Scm shRNA x MTD (positive control
- (all MTD crosses at 30C)
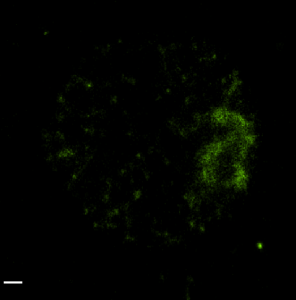
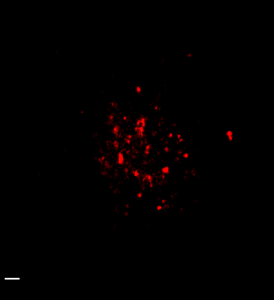
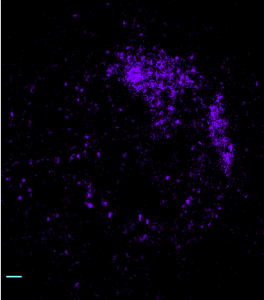
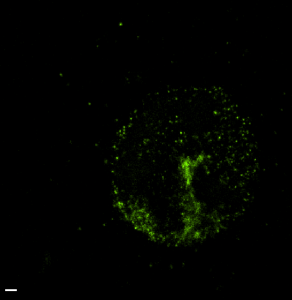
STORM analysis of multi-color HP1 data (2D acquisition and analysis)
(647 green, 750 red, 488 magenta). 750 structure appears substantially undersampled and overly punctate. No general nuclear signal is detected outside of the centromeric DNA zone. (scalebars 500 nm).
Bistability
- writing model section text and figures for manuscript
- retweaked phaseplots of deterministic system