10:30 A – 8:00P
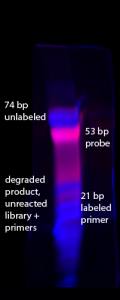
Probe making
snail data analysis
- run nuc mapping cyto2nucmap for MP08Hz_snaD_2 data
- modify lsm_viewer to not append embryo numbers when running export all on embryos data without position data.
STORM
- test out new buffer conditions for 750 dye.
- definitely brighter but substantial reduction in total number of localizations. Not sure if this is fewer molecules responding or fewer cycles.
- 1-3 K frames at 30 Hz and switching is done. (not much active after first few hundred frames)
- dyes are all in off state to start — no conventional imaging.
- compare to 647 labeled HP1 — switches for 150,000 frames (60Hz imaging) at 300 mW illumination with no need for 405 activation. 750 switches for 10,000 in