10:30 A – 7:00P, 8:30P – 9:30P
(8:30 – 10:30 workout session)
Probe making
- combine, precipitate and pellet en-647 oligoPAINT probe
- # Wash, combine, and resupend probe-pellet,
- # Prep for digestion reaction
Cell culture (500 kbp probe test 2)
- After removing probe and adding buffer, sample has just a strong general glow that makes it impossible to make out individual cells. Contrast / individual cells are much easier to see in untreated wells.
- After 15 min 2x SSCT 60C hot wash and buffer change, nuclei are clearly visible from consistent nuclear background. This is substantially more evident in the labeled wells than the non-labeled wells and thus a probable consequence of non-specific probe binding.
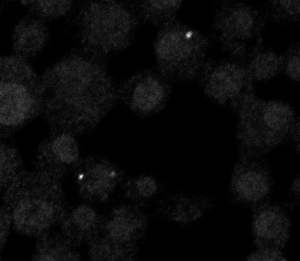
- By confocal, A few cells on the 5:15o dilution of 500 kb probe have signal. Multilayer sheets of cells show no signal — need to plate cells sparser! Some cells show too many spots — background sticking to dying cells? Or 647 background? Untreated wells don’t appear to show this degree of fluorescence so I believe it’s from the added probe.
- Since it’s not 100% of cells, even in the sparse regions, not certain yet to conclude that this probe works.
Coding
- Incorporated DaoSTORM dot finding into STORMfinder
- # Add file and analysis menus to STORMfinder
