Sunday 05/05/13
STORM Analysis
- Fixed bug in RunDotFinder that was causing DaoSTORM analysis on to correctly overwrite/rerun analysis (DaoSTORM method check had method in quotes, causing failure).
Project 2
OligoPaints
- Running 2.5 mL of PCR using PPTC primer at 60C for B,Y,K and G libraries.
- Does not look to good on gel:
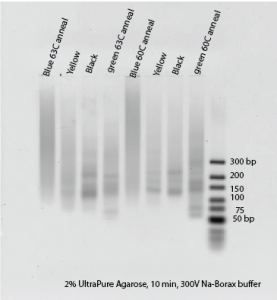
- Blue failed again! This worked so well with inner primers yesterday:
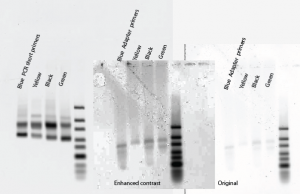
- Set up new PCR gradient 50:70C (12 steps) using PPTC and sublibrary-specific NP primers
- Set up second gradient, added 1.5 uL PCR1 product (amplified with inside primers) and adapter primers + NP primers (+ PPTC from original dilution). This one 55:72C gradient (12 steps), 15 uL reactions.
Qubit
- All measurements in ug/mL
| sample |
PCR |
E1 |
E2 |
E3 |
| Blue |
60 |
46 |
3.6 |
nd |
| K2 |
34 |
31 |
16 |
4.0 |
| AbdA |
15 |
29 |
27 |
15 |
| Ubx |
? |
36 |
37 |
3.5 |
| AbdB |
? |
nd |
nd |
6.3 |
| B 2.5 |
48 |
? |
? |
? |
| Y 2.5 |
25 |
? |
? |
? |
| K 2.5 |
48 |
? |
? |
? |
| G 2.5 |
62 |
? |
? |
? |
Nanodrop
| sample |
E3 |
| K2 |
16 |
| Ubx |
18 |
| AbdA |
46 |
| AbdB |
22 |
Column clean up 2.5 mL scale
- 15 mL (2.5 + 12.5 binding buffer) is just about full volume capacity for the CC-100 columns. (this is true according to spec sheet).
- Spinning at 3,000g with 15 mL on top breaks columns (can be snapped back together)
- Spinning at 300g gets the 15mL through, then need a short spin at 14,000g in mini-centrifuge to elute remaining binding buffer.
- 2x 2mL wash on manifold
- spin dry at 13,000 rpm
- elute with 150 uL 50C ddH2O.
Orders
- # Order more primers
- # Order more probe synthesis reagents + troubleshooting diagnostics.
This entry was posted in
Summaries. Bookmark the
permalink.


