9:30 A – 7:30 P, 8:30 P – 12:30 A
Literature
- Interesting paper by Sanchez and Gore on altruists, cheating, and ecological stability in yeast
- Clear neat article in Nature on being a scientist.
Oligo Paint probes
- Pool O/N PCRs
- Test gel of O/N PCRs on Ubx, AbdA, and AbdB:
*
- AbdB has double band (~200 and ~130). Others have single bands. PCR product seems to like to run slow, ~ 200 bp (effect of proteins?) Everything is in the bouble band state?
- start heating waterbath
- check yesterday’s cast gel — looks okay.
- pre-run large well 1.5mm gel
- cast more large and small Urea PAGE gels.
- Denaturing gel of AbdA, AbdB and Ubx has single bands.

Extracting AbdB and Ubx probes run on 5 well 1.5 mm Urea PAGE gel, ~170 uL per well
- Where did the extra bands come from in the pre-digested gel! Why so many bands in the digested gel!
- Cut out large and small bands

- dissolving at 37C O/N. precipitate and test if these work as probe?
- Ordered new primers (using Brian’s secondaries) for Y,G,B and K.
New PCR, brian’s program
| sample | Elute 1 | Re-elute 1** | Re-elute 2 | fresh E2 |
|---|---|---|---|---|
| Ubx | 275 | 315 | 801 | 223 |
| AbdA | 313 | 348 | 382 | 42 |
| AbdB | 174* | nd | 312 | 87 |
*AbdB is with my homemade binding buffer
**Eluted into discard capsule first, not into collection tube!
T7 try N
- Run PCR of fresh AbdA, AbdB and Ubx on gel
- lower yield using Brian’s PCR protocol than my version with shorter annealing.
- Run second PCR with PPT and PPT control
- 3x large column elution of 1 mL scale PCRs 800, 300, 300 ng/uL yields
- Very low yield:
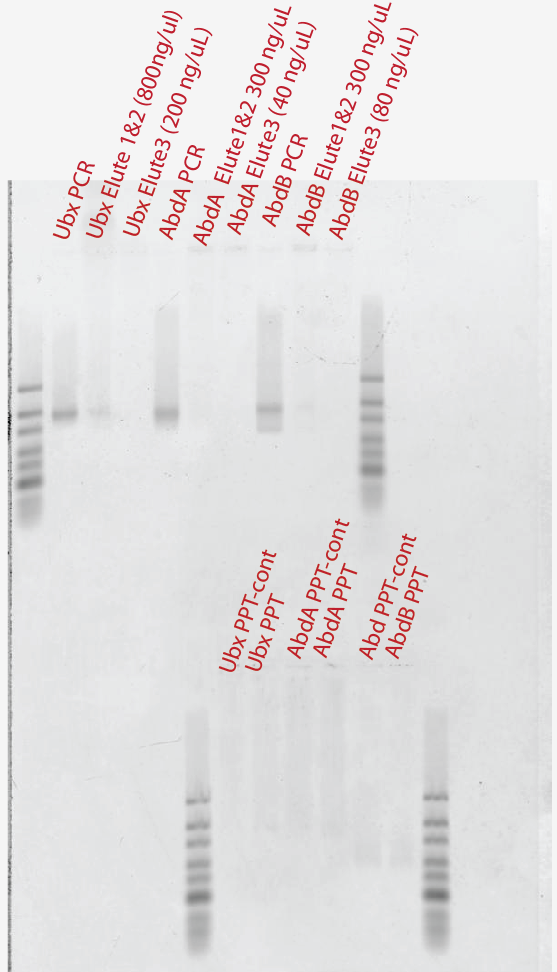
- Looks like both PPT PCR and elutions basically failed. The first elution worked last time at least! Then I started getting extra bands. Time to go back to Glycogen and ethanol percipitation I think.
- Running new PCR at 68 start drop to 62 (for 20mer primers). Hopefully this gives better yield for AbdB and Ubx.
Rewriting Mosaic viewer for new version of Steve
- Trying to get mosaic_to_matlab.py to work on Monet. Needs SIP and or PyQt
- Failed to get sip to install properly on Monet:
Error: Unable to open "C:\Users\Alistair\Desktop\sip-4.14.6\siplib\siplib.sbf" - Failed to get Py QT to install properly:
Error: Make sure you have a working Qt qmake on your PATH. - converter runs on Cajal. Just pass it the
mosaic.mscfile. Pretty slow to run, takes a few hours to convert a mosaic… - Sip did install, it just did it in my matlab-storm working directory rather than C:/Python27 like it said it would. Still not sure if it works, currently converting on Cajal. Very slow…
- Cajal batch run says its runs and converts but I don’t see any matlab files…?
- Writing new mosaic viewer to work with matlab files. No master list to jump to the right files and just load those… :(.
- rewrite not finished.
STORM
- mRNA STORM with Hao’s cells. Switching looks good to me. Try analysis tomorrow
- STORM BrianB sample 4. Essentially no switching. Very low background switching either, though nuclei glow steady under heavy elumination…
- STORM BrianB sample 2. Switching nicely. Running small batch of cells overnight. cell density was better on other coverslips.
