10:00 A – 5:00 P, 9:00 P — 11:45 P
Analysis of signal and background in different hybe conditions
- Discussion with Brian on temperature results
Additional notes not yet sent:
Hybe conditions that reduce binding to reduce background seem to also lose target probes. As the number of probes goes down, one is tempted to image for longer in order to detect a higher fraction of the probes which are bound, and all during this time, waiting for these probes to give their last blinks, we are accumulating background.
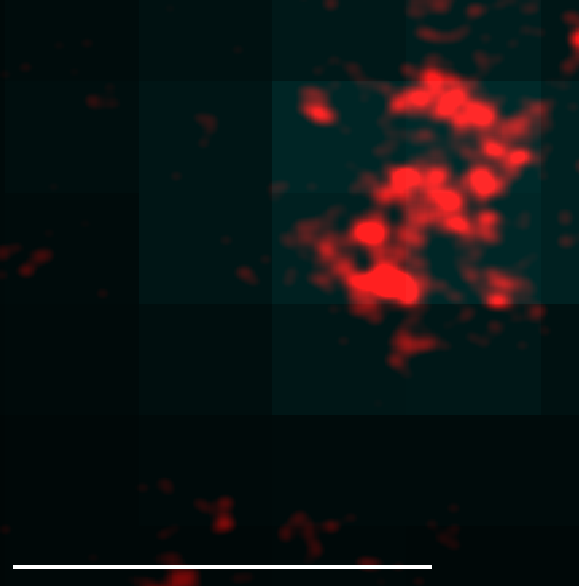
In contrast, if the labeling is really high, we have much more signal in the target spot than in the background spots, so it is easy to identify and in the very small region of the target spot most of the signal will be on target binding events. For example, some of our early BXC images have >5,000 localizations per pixel in the region of the nucleus overlapping the conventional dot, and <200 localizations per pixel on average in the nucleus, which is pretty good signal to noise still (25 fold).
From our preliminary tests with the activator before, I think that may be pretty useful in further reducing background. In particular, if the off target binding events of the 405 labeled or unlabeled primary probe sequence occlude the secondary binding region when they stick to their off target partners, these guys won’t receive cy5 conjugates.
Differences in clone8 vs S2 cell hybes, sent to BB.
Testing biobeads approach for probe making
- see notes
- this post also documents other approaches in the pipeline to try.
Ordering
- Ordered T7 and RecJ
- Carboxyl beads suddenly changed price ridiculously on the Fishcer Sci site.
- Ordered primers for RNA synthesis