10:00 A – 8:00 P, 9:00P – 12:14 A
Feducials
- Suggestions from Ke
- Dual-Objective scope has a dual-pass filter to allow green beads to show through
- Add beads in imaging buffer (no need for washing).
- Try 625 beads.

- Use PBS with Ca[2+], Mg[2+] as imaging buffer.
- Ke uses MEA — fewer photons (~3800 vs 5000) but lower duty cycle (.0005 vs .0012)
- Quick test with 625 beads
- 625 beads may be too bright — need to compare to sample
- Beads also do bleach, not incredibly fast and mostly in a few steps (maybe detach?)
- Beads added in imaging buffer zip around and continue to attach during imaging.
- work out software to do tracking based drift correction.
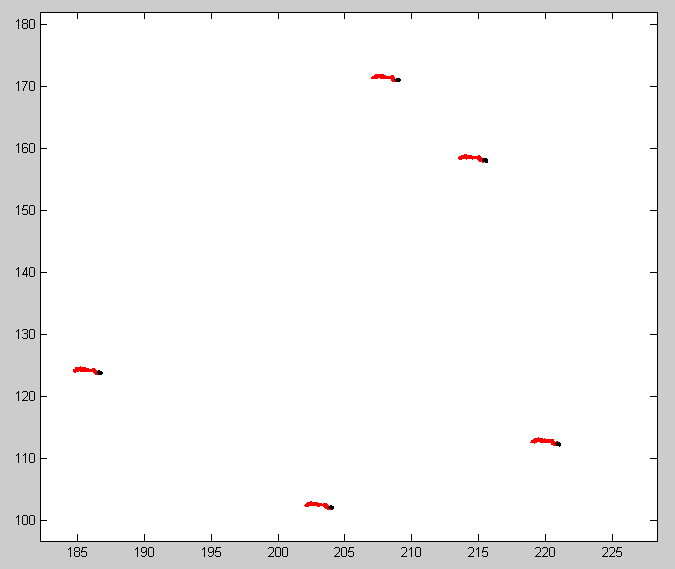
- check color-coded temporal traces of BX-C loci to see if drift is a likely source of dot spreading
- Not obviously extended by drift (not uni-directional at any rate). Not easy to tell though when dots are on for only ~2000 of 60,000 frames unless drift is uni-directional.
More feducial attempts
- 625 beads on embryo coverglass. Don’t see beads, lots of single-molecule red-dye background. Maybe these ancient beads have deterioriated and released dye? don’t have contrast to see beads against bright embryo spots in conv?
- Try 540 beads with weak yellow laser. Easier to distinguish bead-dots from nuclear dots. Don’t bleach.
- imaging AATAT embryo
Probe making
- Restriction enzyme should arrive today.
- Set up nicking reactions
- per 100 uL reaction
- 81 uL DNA,
- 10 uL buffer,
- 9 uL enzyme
- AbdA, AbdB, Ubx: 324uL DNA + 40uL buffer + 36 uL enzyme
- B, G = 162 uL DNA + 20 uL buffer + 18 uL enzyme
- Y, K = 324 uL DNA + 40 uL buffer + 36 uL enzyme
- ND samples before
| sample | ND |
|---|---|
| B EE | 3650 |
| Y EE | 2591 |
| G EE | 4040 |
| AA EE | 1410 |
| AB EE | 567 |
| AB W2 | 45 |
| Ubx W2 | 98 |
- Heating up hot-bath to run gel
- Pre-run pre-cast 15% Urea-PAGE gel, 30 min in hot bath at 55-60C
- Pool digest reactions
- split large reactions into separate 200 uL volumes
- to each 200 uL, add 4 uL glycogen, 25 uL 4M NH4Ac, and 1 mL cold EtOH
- freeze O/N
- Gel does not look promising, only obvious difference is a broad band at <50bp in all the digest conditions. Both pre and post digest have acquired a double band, even though the denaturing gel off the PCR had only 1 band.
- Maybe this will run cleaner after EtOH precipitation
- though wtf what happened to the predigest…
