8:30 A – 5:00 P, 7:30 P – 11:45 P
Probe making
ethanol precipitations
- Ethanol precipitate B/Y/K/G probes from 2-5 mL scale synthesis (which had poor nicking and extra-large bands).
- Ethanol precipitate new 4 mL scale B/Y/K/U PCRs with primer-fixed nick sites
- low/no glycogen large scale percipitates, with substantial protein component, the pellet fragments easily and it is difficult not to lose bits of pellet when removing the EtOH.
- resuspend probe DNA in hybe buffer. Measure concentrations:
- B/Y/K ~ 300 ng/uL, gel-purified ssDNA, in ~50 uL hybe buffer = total yield (measured on nano-drop).
- G ~ 30 ng/uL ssDNDA in 50 uL hybe buffer
- concentration ~ 300*3.3/96 ~ 10 pmol per uL
- previous en stain was at 150 ng/uL final concentration (in 25 uL that’s 250 pmol).
- resuspend PCR EtOH precipitate in 320 uL, vortex. Additional flasks are the the Y and K 2 mL scale reactions that failed to amplify well.
Nicking reaction
- ~300 uL recovered, resuspended product. put aside 4 uL diluted into 18 uL TBE for test gels etc.
- to remainder, add 38 uL 10x ‘CutSmart’ buffer
- add 35 uL Nicking enzyme Nb.BsrDI
- split into 7.5 50 uL reactions in PCR strips, react 5 hrs, heat denature, fridge (run in thermocycler O/N).
Test staining:
- Poly-lysine treat 6 22mm 1.5 glass coverslips
- add Kc167 cells, allowed ~4 hours to settle and attach
- cells perfectly trace out where poly-lysine sat on coverglass
- sodium borohydride teat cells, 5 min (little over on the concentration)
| sample: | Blue probe | Yellow probe | Black probe | Ubx probe | en-cy5 primary | no primary |
|---|---|---|---|---|---|---|
| primary | 8ul = 80 pmol | 8ul = 80 pmol | 8ul = 80 pmol | 8ul = 80 pmol | 80 pmol | 0 |
| 2ndary | 5 pmol | 5 pmol | 5 pmol | 5 pmol | 0 | 5 pmol |
- Over-diluted secondary: currently at ~1 pmol/uL. Should have ordered this at a larger scale anyway.
- denatured on downstairs heat block at 91C, 3 at a time, starting with the first 3.
- incubating in humidity chambers at 40C O/N upstairs.
T7 polymerase production of RNA probe:
Nanodrop concentration
- T7 P1: 106 ng/uL – used 10 uL for T7 Pol II reaction (~ 1-2 uL left)
- T7 P1rc: 113 ng/uL – used 10 uL for T7 reaction (~ 1-2 uL left)
- Ubx-adapter emPCR elute 2: 8.2 (may be all background) = tossed
- B-adapter emPCR elute 2: 5 (may be all background) = tossed
- K1-adapter emPCR elute 2: 57
- Ya-adapter emPCR 9.4 (may be all background) = tossed
NEB
- High-Yield T7 protocol
- recommends 4-16 hrs for template < .3 kb.
- Set up reaction at 20 uL scale according to prtocol for both primary and rc-primary T7 Blue library.
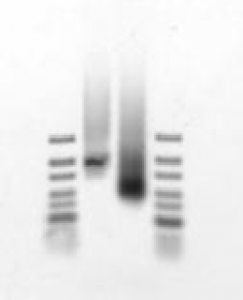
- At 8 hrs, running out small aliquot of reaction on agarose gel. sample 1 (no primary no rc) and sample 2 (rc-primary)
Promega
- Transcription Optimized 5X Buffer 20μl
- DTT, 100mM 10μl
- Recombinant RNasin® Ribonuclease Inhibitor(a) 100 units, 2μl
- rNTP mix (2.5 mM of each) (see Section II) 20μl
- DNA template, linearized (in water or TE buffer at 2–5μg) 10 uL
- Phage RNA Polymerase 40 units (~80 U/uL) 1μl
- RNAse free ddH2O 45 uL
STORM
- finish O/N imaging of 488-wt-ph 647-ph-flag
- shoot 3D bead fields for chromatic alignment
- Imaging project 2 cells with Hao and Steven
- set up O/N imaging of 488-wt-ph, 647-phM-flag
