9:30 A – 7:45 P, 8:30 P – 12:00 A
expanding primer sets
- looking into quaternary cyclic codes to generate primers with appropriate distance
- There is literature on this, but it is not obvious from the proofs and coding theory descriptions how to implement the code.
‘quicker’ straightforward approach
- using 25mer primers as a base set of sequences from Elledge lab
- converting into 20mer primers with GC clamp and Tm filter. Current parameters leaves ~80K of 240K (Tm 65-75 still a bit high for 20mers, I think the 70-80 Tm of the Jeff set also substantially limits library size).
- raised Tm to the 70-80, kept 29,329 primers. This will probably still take the rest of the day and maybe night to BLAST against mouse…
- Test run with first 10000 BLAST against genome: 489 have 16 or fewer hits to mouse
- Fixed up a few code errors, hopefully running nicely now O/N on Mouse, Fly, Human, Monkey and E. Coli genomes.
Ordering
- ordered T7 NEB kit
- ordered aminoallyl UTP
- ordering T7 primer adapters for BX-C components
probe making
- use Zymo kit from Jeff to Gel extract
- looks much better (first 2 columns P1, RC, before gel extract, second two, RC, P1 after gel extract)
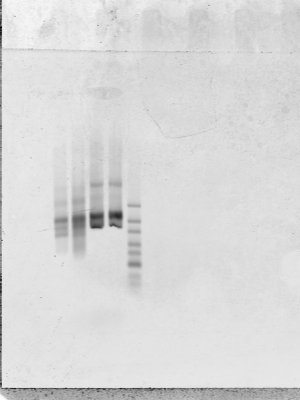
- still some large band — seems to be a denature/renature biproduct of the smaller bands (which is also why we get two bands again, though I just cut one).
- Got T7 high RNA NEB kit from Jeff, ordered new second generation recently release kit for Jeff to replace
- 4.5 uL of ATP, TTP, and GTP, 3 uL of CTP plus 20 uL of CTP-cy5 = maser mix (that’s basically all my CTP-cy5).
- 100 nmol CTP is only like 3 reactions!
- running O/N 16 hr tx reactions with 1 uL RNasin added and 10 uL new-made CTP-cy5 RNA mix.
STORM
- Teaching Guisheng and Jiang about multicolor imaging with Steve and Dave software on STORM 2 and STORM 2 protocols.
- set up O/N STORM on new sample round 2 (well really round 3), slide 1.
- very low background, bright dots, not much switching though. probably labeled secondary no primary.
- setup complete imaging session 2x
- setup O/N image analysis of Gdc2 data.