10:10a – 7:00p, 8:50p – 12:10a
STORM & staining BX-C tiles
AbdA labeled coverslip (S2 cells)
- found cross-hatching
- imaging additional cells
- record chromatic beads
- will restain AbdA-stained coverslip with BX-C
Ubx labeled coverslip (S2 cells)
- found cross-hatch (much more evident with visible light in 647 QV channel.
- added 2 mM (final) COT to buffer.
- ubx dots much brighter, much more evident per cell.
- excellent contrast, excellent single molecule brightness, excellent switching, excellent 405 responsiveness.
- recording 95K frames in 256 x 512 mode.
Useful notes
- Hazen and Graham have screened COT properties. see email.
project 2
- helping Hao get set up with PCR steps
- PCRs look okay, very low nanodrop yields
Cell culture
- passaged Kc cells: attempted to remove floating cells and leave attached ones in dish. Added new media
- Biked to Med school to pick up clone 8 cells
- used improvised scraper to passage clone 8 cells (easier than trypsin, check tomorrow if this worked).
Probe making
- primer plates arrived
- don’t have an unlabeled short common (or labeled or secondary tagged common). Will have to wait till next week to start.
- picked up BX-C primaries and secondaries from Bryan.
- Requested to test Tye-6?? dye as a cy5 substitute for STORM
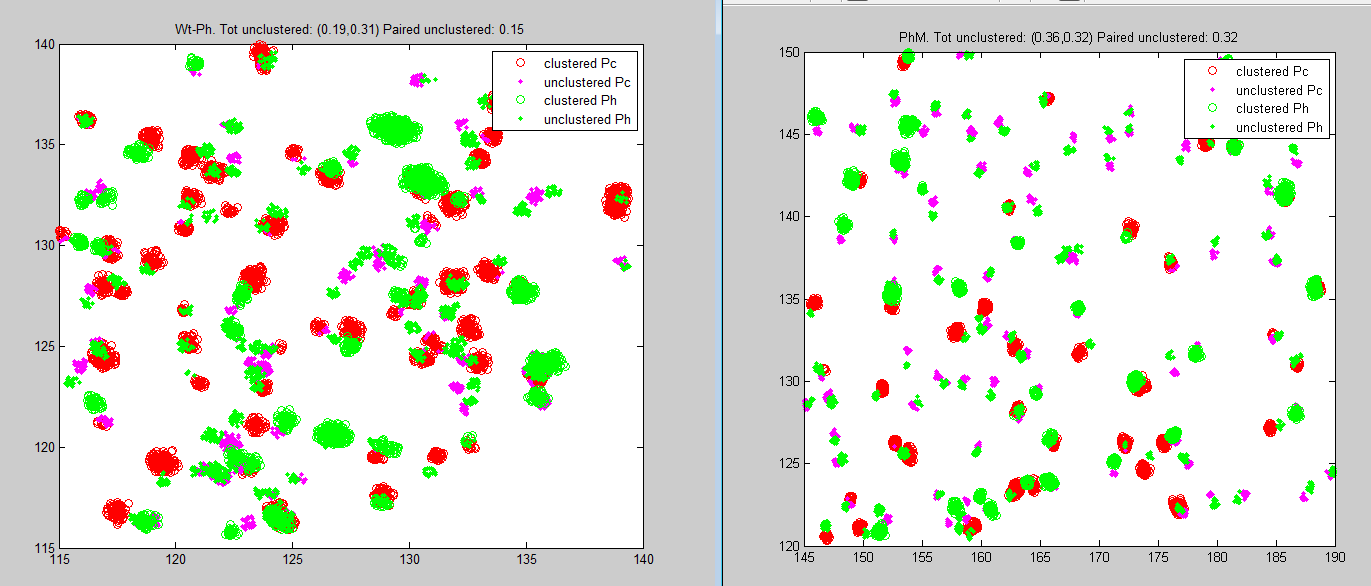
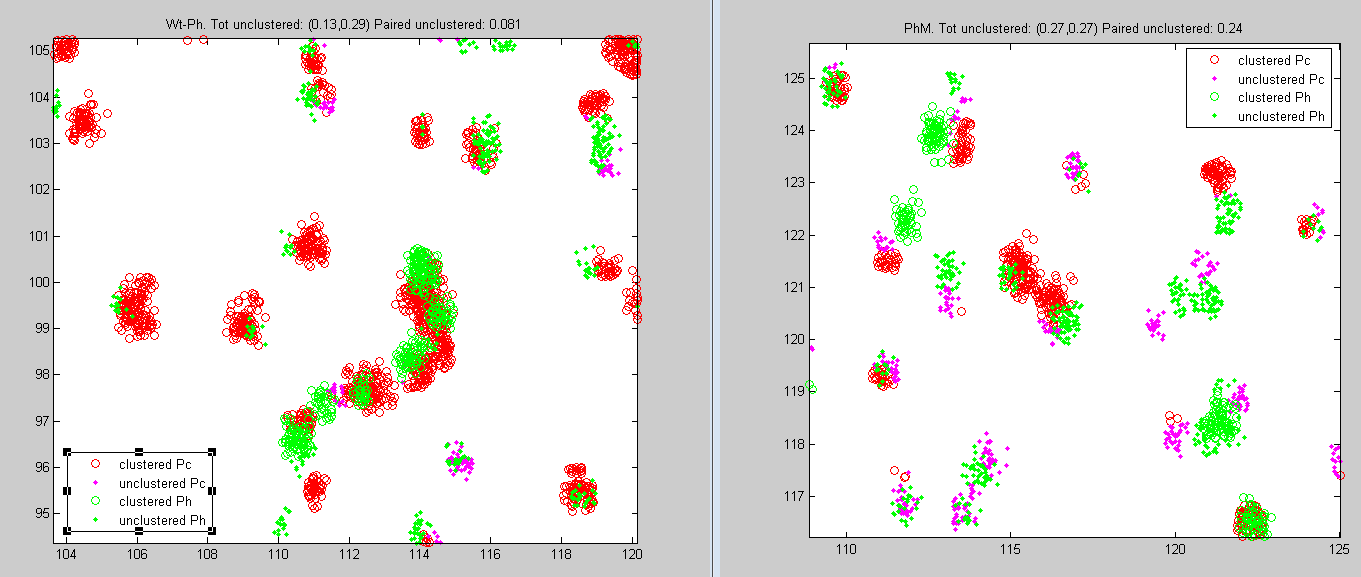
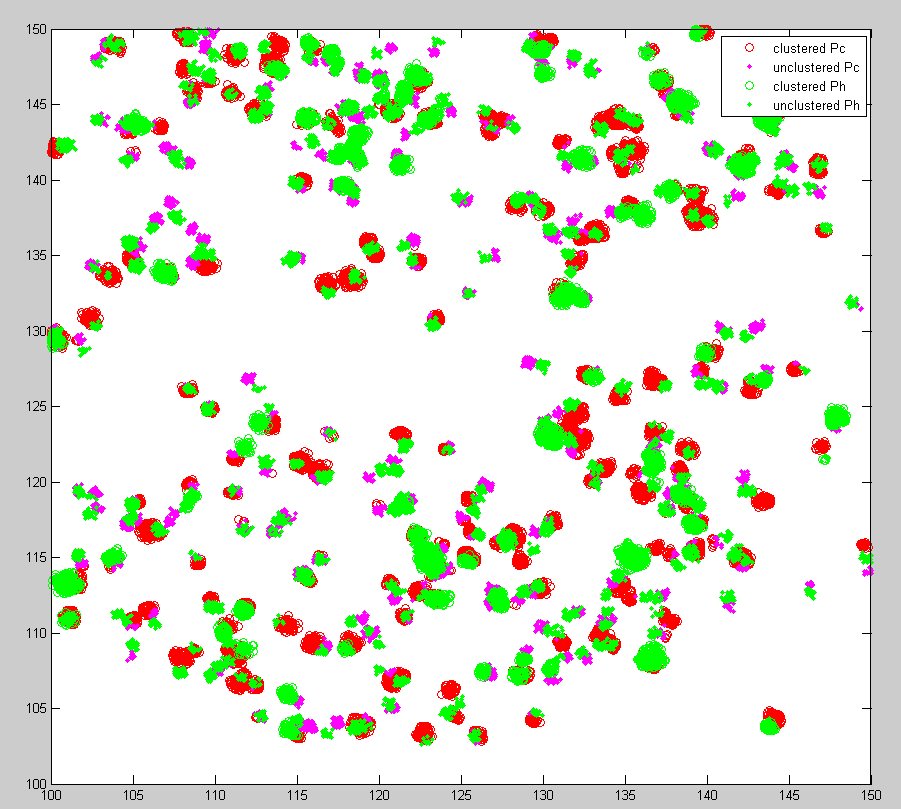
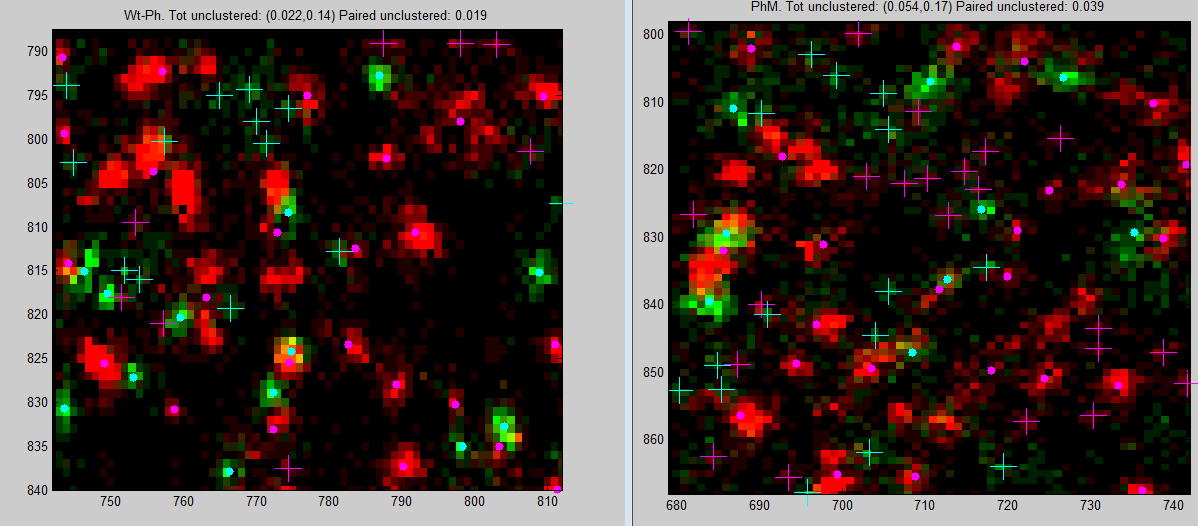
Ph data analysis
Cell labeling
- relabel AbdA coverslip with Ubx: 3 uL Ubx-primary from 10ug-prep, + 1.5 uL secondary + 1 uL new 1:20 diluted 405 tertiary. forgot RNase, (hopefully not needed?)
Review feedback from Jeff
- Be sure to walk reader through each panel of a figure in the text.
- Make a clearer link between the ‘cool new tools / things we can measure’ and how these relate to model predictions.
- don’t worry about length
- Distinction between Sanchez approach and Evans approach is confusing. Draw picture:
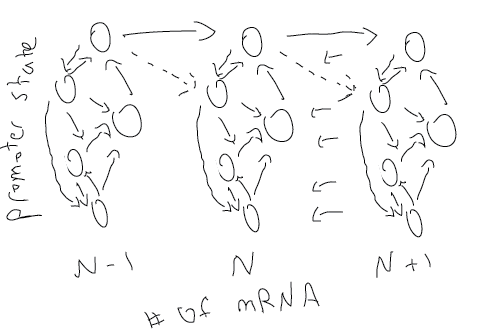
- Talk about variability in time not variability in rate.
- Make it clear when you add degradation and when you don’t.