9:30a – 11:15p
STORM
- stop O/N STORM
- start copying data to Monet from O/N
- start spot-finding analysis on yesterday’s O/N splitdax F6 data.
- G2 data also not analyzed: set up DaoSTORM analysis on Cajal
- G2 bead data looks sparse — should check a few late samples and consider deleting the 561 splitdax channel to save disk space.
Probe Making
Library quality check
- Finish design and order of deep sequence adapter primers for library
- discuss design with Jeff
- recommendation: 96 well amplification PCR, take 1 uL of each and pool these together and then run adapter PCR on that.
- also take some of the independent samples directly and run separate PCRs on each.
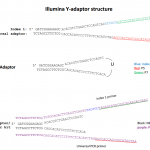
Reverse transcriptase reactions.
- take stock of RNA in 100 uL format not made into probe
Probes to try from plate (~100 kb+)
- D11 100 kb percicentric Green
- E01 200 kb Green
- E8 75kb Red
- E10 98 kb pure yellow
- E12 630 kb Green
- F1 BXC_K_left, 125kb pure K
- F3 Ubx through bxd
- F4 AbdA through iab4
- F5 iab5 through AbdB
- F7 Bxc_K right 240 kb solid K
- F10 478 kb Black
- F11 ANTCleftBlack 180 kb
- G9 76 kb Blue near ANTC
- G10 425 kb black region
* all F probes have insufficient volume (e.g. BX-C genes F3 – F7).
* very low yield from 13 uL of RNA isolated on beads and resuspended in 40 uL of ddH2O
* nanodrop: 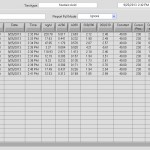
PCR
- D11 100 kb percicentric Green
- D12 385 kb Yellow
- E01 200 kb Green
- E8 75kb Red
- E10 98 kb pure yellow
- E12 630 kb Green
- F1 BXC_K_left, 125kb pure K
- F3 Ubx through bxd
- F4 AbdA through iab4
- F5 iab5 through AbdB
- F7 Bxc_K right 240 kb solid K
- F10 478 kb Black
- F11 ANTCleftBlack 180 kb
- G9 76 kb Blue near ANTC
- G10 425 kb black region
- Neg control 1
PCR short-regions with short primers
- E03 BLUE 13 KB (no genes, good PC/Psc)
- E04 BLUE 10 Kb (flanked by strong PolII)
- F08 RED 7 kb region next to black 1, tRNA locus
- G07 AlphaTub84 (5 kb yellow)
- G01
- G02 (positive control)
- Neg control 1
- Neg control 2
Short-regions RNA clean-up, re-numbered:
- (1) E02 13 kb green (between R/Y)
- (2) E06 Epc (12 kb YELLOW)
- (3) E07 Tou (15 kb YELLOW)
- (4) E09 RED 8 kb intronic tRNA locus, in between blue
- (5) F02 BXC_Y_left (15 kb)
- (6) F09 BLACK 18 kb region next to RED1
- (7) F12 Taf1_(17kb yellow/green)
- (8) G01 LabRegion_(Lab_Zen2) [83 kb but missing piece of ANT-C]
- All concentrations in the 1500 – 3000 (mostly ~2500) ng/uL of RNA in 40 uL.
Short-regions RT reactions: 20 uL scale
- 5uL of ~2500 ng/uL RNA. ~10 – 12 ug of RNA.
- 80*5 – 400 pmol RNA. add 300 pmol of primer.
- 5 uL RNA + 3 uL Primer + 1.5 uL dNTPs + 3.5 ddH2O
- 9x primer master = 27 Primer + 13.5 dNTPs + 18 ddH2O (8 each)
- Raw RNA (3 uL) + 3 uL primer + 2 uL dNTPs + 5 ddH2O
- 9x primer low conc master = 13.5 primer, 13.5 dNTPs, 36 uL ddH2O (7 each)
- per reaction (1 uL SSIII + 1 uL RNasin + 1 uL DTT + 4 uL FSS buffer)
- 18x master mix = 18 + 18 + 18 + 72 FSS buffer (7 each)
Prep
- Clean all RNA columns: 1x wash in .1 M Hcl (400 uL). 5x wash 1 mL H2O. 2x wash in binding buffer.
Black regions (Bogdan).
- RNA clean up. Concentration results below.
- Set up RT reactions, 40 uL scale 1.5 hours, ratio of 150 pmol primer per 5ug of RNA.
| Name | ng/uL | A260 | A280 | 260/280 | 260/230 |
|---|---|---|---|---|---|
| RnaC | 112.47 | 2.812 | 1.243 | 2.26 | 1.19 |
| Rna1 | 158.51 | 3.963 | 1.969 | 2.01 | 2.10 |
| Rna2 | 197.73 | 4.943 | 2.491 | 1.98 | 2.16 |
| Rna5 | 355.70 | 8.893 | 4.354 | 2.04 | 2.34 |
| Rna7 | 413.17 | 10.329 | 5.087 | 2.03 | 2.36 |
| Rna8 | 193.22 | 4.831 | 2.478 | 1.95 | 1.70 |
| Rna9 | 568.47 | 14.212 | 6.669 | 2.13 | 2.10 |
Fly work
- flipped fly stocks that didn’t get flipped on 8/26. Other stocks look pretty healthy (and current food is from 8/28). Maybe flip in a two weeks or so.
