10:00a – 10:40p
Goals
- Laundry
- email Ting & Co. draft letter on progress on goals on chromatin
- Analyze existing data for new spots: density, puncta etc. scale factors? (density vs. length or something?)
Computer
- splitdax aborted in night when ProRAID ran out of disk space.
- shuffling all embryo data to Alistair10.
- Running STORM analysis on F6-405 data. Some movies launch and immediately exit and I’m not sure why…
- attempting to order new Mediasonic 8 bay harddrive enclosure with 2 4Tb Red drives. (Try the add drives as we go model rather than have one big expense. Also hard-drives seem to fluctuate in price a lot and on average decrease).
Probe making
large regions reloaded:
- Test gel for new PCRs
- D11 100 kb percicentric Green
- D12 385 kb Yellow
- E01 200 kb Green
- E8 75kb Red
- E10 98 kb pure yellow
- E12 630 kb Green
- F1 BXC_K_left, 125kb pure K
- F3 Ubx through bxd
- F4 AbdA through iab4
- F5 iab5 through AbdB
- F7 Bxc_K right 240 kb solid K
- F10 478 kb Black
- F11 ANTCleftBlack 180 kb
- G9 76 kb Blue near ANTC
- G10 425 kb black region
- Neg control 1
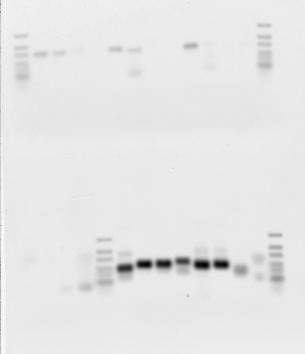
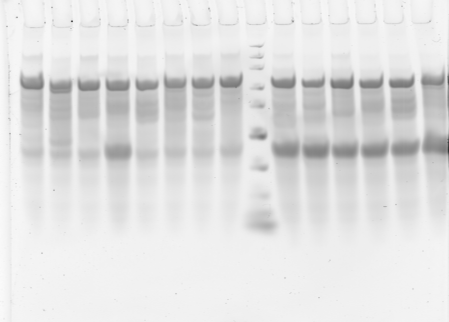
Gel 1 annotations:
- Loaded 1-16 top to bottom right to left, then 1-8 of 50 uL scale reactions with short primers. 2 uL PCR mix per lane.
- Very poor results. Try again with first 6, 50 uL scale.
- 50uL scale reactions with short primers worked much better.
- set up new 50 uL scale PCR with T7 extensions and .5 uL of round 1 PCR for 2-step reactions. used 2.5 uL of previous 1:20 dilutions of T7 primers for short stuff. + Neg controls 1 and 2 (abdA-NiP and abdB-NiP)
- Set up new 50 uL scale PCR reactions for large regions 1-5 + Neg control 1 (ubx-NiP)
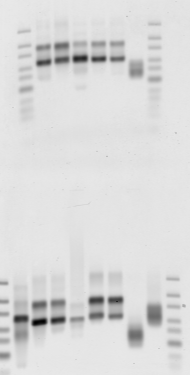
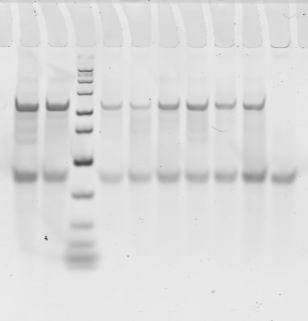
Gel 2
- Top row: Large regions 1-5. Bottom row. small regions:
- E03 BLUE 13 KB (no genes, good PC/Psc)
- E04 BLUE 10 Kb (flanked by strong PolII)
- F08 RED 7 kb region next to black 1, tRNA locus
- G07 AlphaTub84 (5 kb yellow)
- G01
- G02 (positive control)
- Neg control 1
- Neg control 2
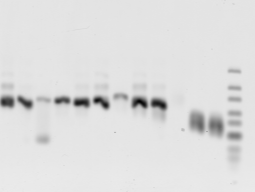
gel 3 annotations
- Tablet driver stopped responding again until computer reboot.
- new PCR of 6-15 (E12, F1, F3, F4, F5, F7, F10, F11, G9, G10). G10 failed. Neg controls do amplify junk again. 🙁 Fortunately this has a different size / appearance and less intensity than what’s observed in the sample channels.
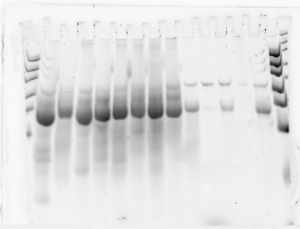
RNA QC:
- Set up denaturing gel (~2 months old? qualitative inspection of gel looks good though).
- Running small regions RNA samples 1-8 + low yield RNA bead cleanup of large regions 1-5 + ladders on each end (15 lanes).
- Gel results:
RT QC and Purification
- small regions
- Bodgan Black regions
- Pour 4 new 15% PAGE gels.
- Run PAGE gels
- Gel1: small regions Raw RNA samples (1-8), DNA ladder, small regions Pure RNA samples (1-6),
- Gel2: small regions pure RNA samples 7-8, Black regions 1-7.
- Black black domain IDs
- 10 kb black domain: D09, next to Red3
- 10 kb black domain: F09, next to RED1
- 50 kb black domain: G08, between GRN and ANTC
- 50 kb black domain: E05, upstream of E(Pc)
- 100 kb black domain: F01, BXC_K_left, 125kb,
- 250 kb black domain: F07, BXC_K_right, 240kb,
- 250 kb black domain: F11 ANTC-L 180kb
- 500 kb black domain: F10, 478kb region, on 3R.
- 500 kb black domain: G10, 425 kb on X
- AbdA-T7 negative control
Writing
- Finish first pass through trimming revision of Tx Review
- Currently 45,000 characters, down from 55,000 characters. 12.5 pages instead of 15.25 (Including figures, not including coverpage/abstract/references)