10:00a – 11:30p
Goals today
- repeat plate PCR, work on technique, set up for sequencing
- STORM pipeline, G10 425kb black. start new small black.
- work on Banff Report – NO PROGRESS
- work on slides for XZ meeting – [Full time spent processing images]
Notes for Bogdan
- 50 uL PCR
- Try non-T7 primers for wells that don’t work.
- Add Dm01-488 stain to cells.
STORM
- Add Dm01-488 stain to cells.
Data analysis
- G2 beads not yet analyzed, running now.
Probe sequence varification
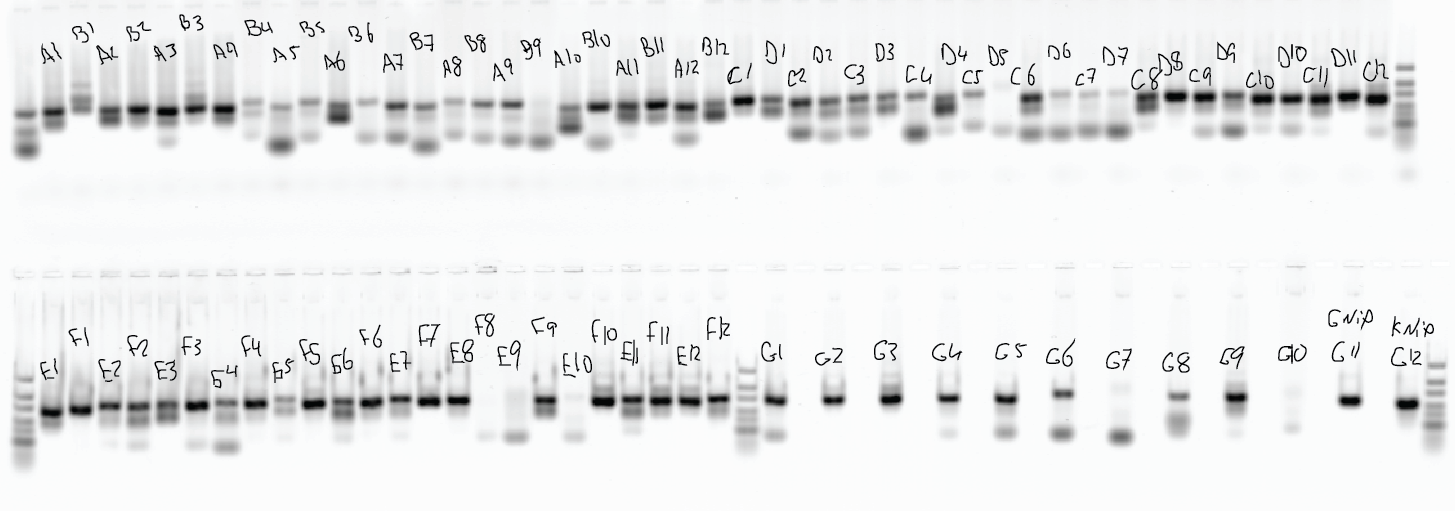
- Plate PCR, 25 uL scale
- Each well:
- 10 uL 2x Phusion
- 12.5 uL ddH2O
- 1.25 uL 10 uM common
- 1.25 uL 10 uM specific
- 1 uL 1:20 dilution library
- Master Mix (90x for 82 wells + 2 nc)
- 900 uL 2x Phusion (450 per tube)
- 1125 uL ddH2O (562.5 per tube)
- 112.5 uL 10 uM common (56.25 per tube)
- 900 uL 1:20 uL library (450 per tube)
- neg controls = Green NiP and Black NiP.
- Pour 150 mL of fresh 2% sodium borax ultra-pure agarose gel with 2x lanes of 75mm wells
- Not all wells look good (see gel below), negative controls have bands.
- sequence, see what comes with the negative controls, see if we get any representation from the libraries without bands.
- Gel:
PCR to add NEB adaptors
- Samples: A1, C5, E6, F3, F5, G3, E2, F2, pool1, pool2 (2 uL of each)
- dilute 1:10 each adaptor sequence.
- to each: add 1.25 uL T7-adaptor, 1.25 uL common-adaptor, 12.5 uL 2x Phusion, 12.5 uL ddH2O
- 11x master = 13.75 T7-adaptor, 13.75 ul common, 137.5 uL Phusion, 137.5 uL ddH2O, 23 uL each.
- 30 cycles NEB primers added (should have cut to like 10 cycles with this much template)
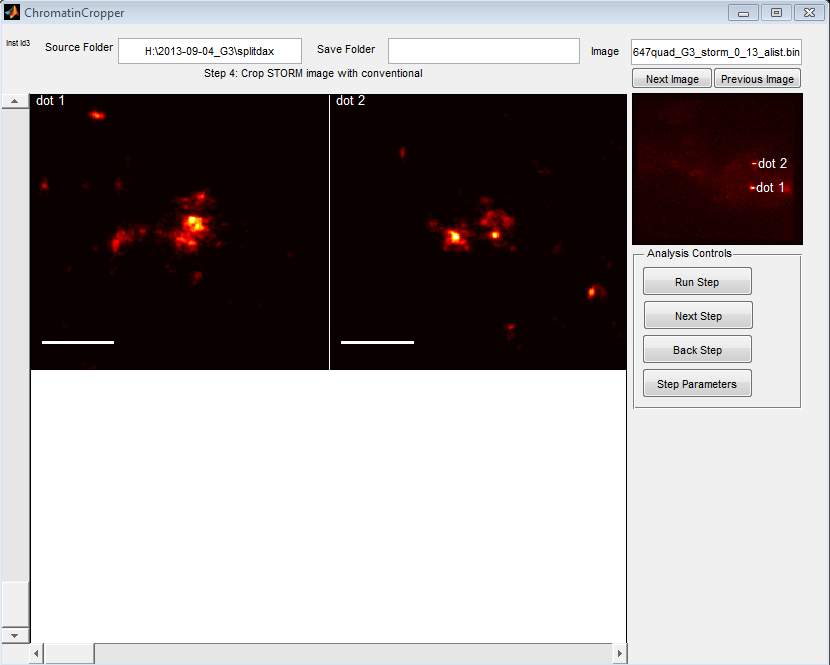
Coding chromatin region analysis GUI
Goals
- parse all conventional images to ID spots. Add filter to exclude out of focus spots and junk.
- extract spot image and molecule lists corresponding to conventional spots
- save
- spot conventional image,
- spot STORM image
- spot molecule list
- spot summary statistics (density, density regions)
Steps
- Load conventional image
- extract spot from conventional image
- Load STORM data
- Drift correct data, use conventional as filter to ID spots. Print a bunch of image views
- save data for selected dots
Initial implementation
Project2
- helping Hao with matlab coding to cuts on regionprops
- working out 6 bit experiments
STORM imaging
- Imaging sample D09
- spots look good. 100% of cells have spots (size 18kb). Let’s see if we can get this size lower. Not bad for a non-transcribed region.
- also imaging Dm01-488 (direct label). Conventional looks good. STORM looks like nothing at all (not needed though).