10:40a – 12:00a
Deep seq lib test
- gel clean up.
- 12 samples x 4 dilutions x 3 replicates = 144 samples
- dilution buffer = 10 mM Tris pH 8 + .05% Tween-20 (500 uL 1M Tris HCl + 250 10% Tween 20 + 49.25 mL ddH2O)
- dilute original library 1:100.
- do the serial dilutions in a 96 well plate and transfer to the 384 well plate. 1 uL 99 dilution buffer.
- 10 uL reaction volumes = 4 uL of template reaction + 6 uL of qPCR/Prmer mix
- Run qPCR on 384 well plate
- should have done stronger dilutions, almost all samples outside of standard curve.
Layout for 384 well PCR
| 1 | 2 | 3 | 4 | 5 | 6 | … |
|---|---|---|---|---|---|---|
| 1 | 1 1:4 | 2 | 2 1:4 | 3 | 3 1:4 | … |
| 1 | 1 1:4 | 2 | 2 1:4 | 3 | 3 1:4 | … |
| 1 | 1 1:4 | 2 | 2 1:4 | 3 | 3 1:4 | … |
| 1 1:16 | 1 1:64 | 2 1:16 | 2 1:64 | 3 1:16 | 3 1:64 | … |
| 1 1:16 | 1 1:64 | 2 1:16 | 2 1:64 | 3 1:16 | 3 1:64 | … |
| 1 1:16 | 1 1:64 | 2 1:16 | 2 1:64 | 3 1:16 | 3 1:64 | … |
| 1 1:256 | e | 2 1:256 | e | 3 1:256 | e | … |
| 1 1:256 | e | 2 1:256 | e | 3 1:256 | e | … |
| 1 1:256 | e | 2 1:256 | e | 3 1:256 | e | … |
| 1 1:1024 | e | 2 1:1024 | e | 3 1:1024 | e | … |
| 1 1:1024 | e | 2 1:1024 | e | 3 1:1024 | e | … |
| 1 1:1024 | e | 2 1:1024 | e | 3 1:1024 | e | … |
| S1 | S2 | S3 | S4 | S5 | S6 | e |
| S1 | S2 | S3 | S4 | S5 | S6 | e |
| S1 | S2 | S3 | S4 | S5 | S6 | e |
Probe making
- PCR cleanup
- set up T7 reactions
Mentoring
- Sample order confused, Bogdan repeating RT reactions
- Need chromatic correction to align 488 nuclear envelope images!
- Running chromatic data from last Ph dataset (still haven’t analyzed this data set).
Ph project
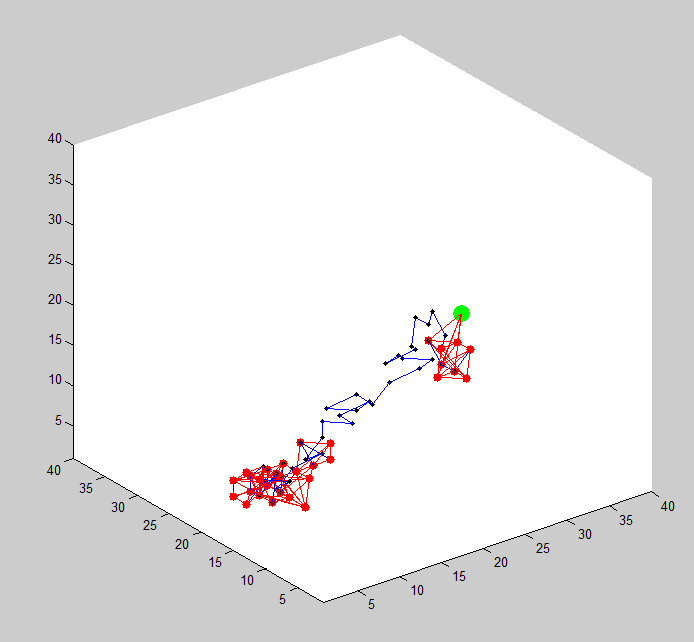
- Generalize random walk polymer to 3D. Success!
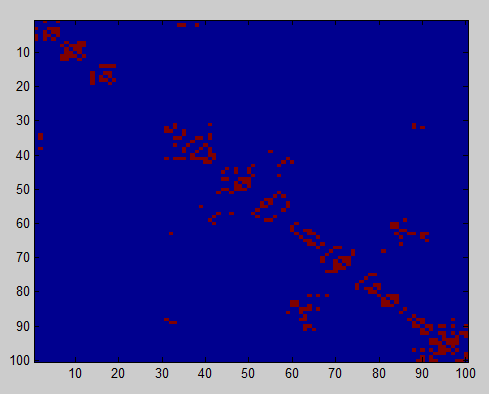
- Compute all neighbor collisions
- Attempt to make joined nodes move as a block. (otherwise these regions won’t find eachother).
- simulations seem to be working!
T7 reactions
- 8 uL rNTPs, 2 uL T7, 1.5 uL 10x buffer, 8.5 uL template
- 9x Master mix: 18 uL rATP, 18 uL rUTP, 18 uL rGTP, 18 uL rCTP, 13.5 uL 10x buffer, 18 uL T7
- reacting O/N (stated ~8pm)