8:30a – 10:30p
Goals
- Ph simulations: time and optimize?
- Ph simulations: add Ph-Flag species
- Analyze qPCR data
- chromatic warp computations for Bogdan
- Banff report
- RNA prep/ clean up
- RT reaction
- cast gels with Bogdan
- test RT reaction
qPCR analysis for lib prep
- Matlab can’t read new xlsx format BioRAD data (despite it’s claims to the contrary):
- Excel freezes horribly on Cajal while trying to save data as regular excel, no amount of End task / End process will close it.
- see Matlab script
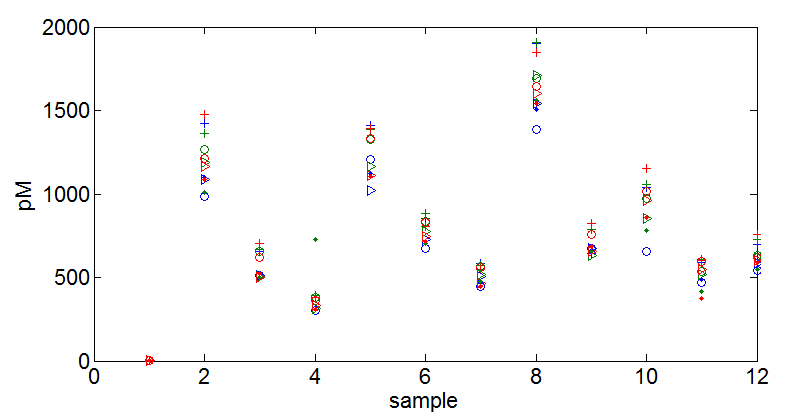
qPCRLib2Prep_131021.m - colors are replicates. symbols are different dilutions
- 1x dilution and 1024x dilution a bit outlier, excluded. (1x way off calibration scale too).
- 256 ‘+’, 64 ‘0’, 16 ‘>’, 4 ‘.’
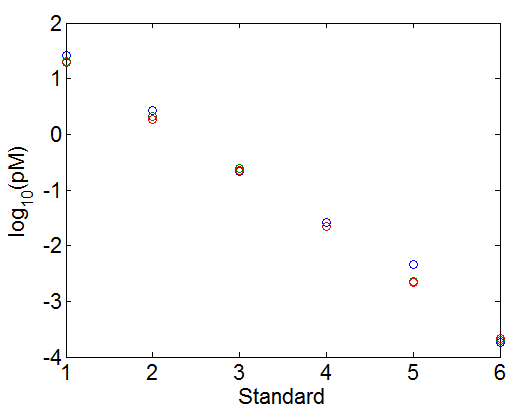
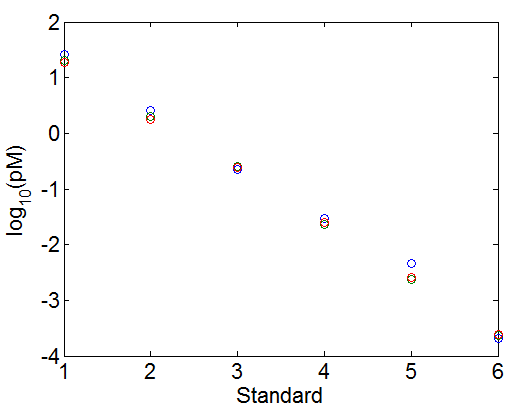
concentration calculations v2
Probe Making
RNA clean up and Quant
- RNA cleanup sample #7 (G9) and #8 (T7 positive cntrl)
- #7 = 1890 ng/uL from 10 uL T7-RNA solution eluted in 30 uL of ddH2O.
- raw T7 solution is 3x more concentrate ~6000 ng/uL ~6 ug/uL
RT reaction
Test Maxima concentration:
- 3 uL G9 clean + 1 uL cy3 + 2 uL dNTPs + 9.5 uL ddH2O
- 6x master = 18 uL G9 clean + 6 uL cy3 + 12 uL dNTPs + 57 uL ddH2O.
- Add 15.5 uL to each of 5 tubes.
- 1 uL Maxima, .5 uL Maxima, .25 uL Maxima, .125 uL Maxima, 0.0625 uL Maxima
- Maxima setup: serial dilutions in 5x-buffer 2 uL Maxima + 2 uL 5x-buffer. Take 2 uL add to 2uL 5x-buffer.
- To dNTP/primer/RNA mix, add 2 uL 5x buffer + 2 uL diluted Maxima + 0.5 uL RNase-inhibitor
- buffer RNase-inhibitor mix = 7*2 + 3.5
Test for high yield
- 10 uL RNA #1 (~60 ug) + 10 uL cy3 primer + 6 uL dNTPs (26 each)
- 10 uL RNA #2 (~60 ug) + 8 uL cy3 primer + 6 uL dNTPs + 2 uL ddH2O
- 10 uL RNA #3 (~60 ug) + 6 uL cy3 primer + 6 uL dNTPs
- 10 uL RNA #4 (~60 ug) + 4 uL cy3 primer + 6 uL dNTPs
- 10 uL RNA #5 (~60 ug) + 10 uL cy3 primer + 6 uL dNTPs
- 10 uL RNA #6 (~60 ug) + 6 uL cy3 primer + 6 uL dNTPs
- 8 uL buffer, 2 uL RT, 1 uL RNasin, 3 uL ddH2O
- 56 uL buffer, 14 uL RT, 7 uL RNasin, 21 uL
Probe making
- 10 uL RNA #1 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs 24 uL
- 10 uL RNA #2 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs
- 10 uL RNA #3 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs
- 10 uL RNA #4 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs
- 10 uL RNA #5 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs
- 10 uL RNA #6 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs
- 10 uL RNA #7 (~60 ug) + 8 uL 405 primer + 6 uL dNTPs
- 8 uL buffer, 2 uL RT, 1 uL RNasin, 5 uL ddH2O
- 64 uL buffer, 16 uL RT, 8 uL RNasin, 40 uL ddH2O
Mentoring
- Bogdan having issues casting gels. May have been issue mixing APS and TEMED together prior to adding.
- Cast new gels. One broken piece of glass (knicked at the bottom) caused solution to all leak out. Other 3 gels will hopefully cast alright
- Not happy with chromewarps. Needs some optimization. Zoom in before and after images correspondence not obvious — more localizations in after warp? Draw lines shwoing who’s matched to who?
- Fixed analysis (needed offset of >15 (20 works) for original bead match and search radius >2 (6 works) for polynomial fit. Then sufficient beads were found. These warps do line up the beads rather nicely. Maybe I should make chromewarps save the image file of superimposed beads rather than the dot plots…?