9:15a –
Deep seq prep / discussion
- Miseq = 25M reads. Have almost 92K unique sequences in my library, so need 1M sequences just for 10 fold coverage.
- recommend run all libraries at equal concentration — we’ll get a lot more sequences from the specific libraries
Sample composition
- Whole library (91K sequences), independent replicate 1.
- Whole library, independent replicate 2.
- Whole library replicate 2 with different index.
- 9 variable size individual sub-libraries (100 – 10K members)
Probe Making
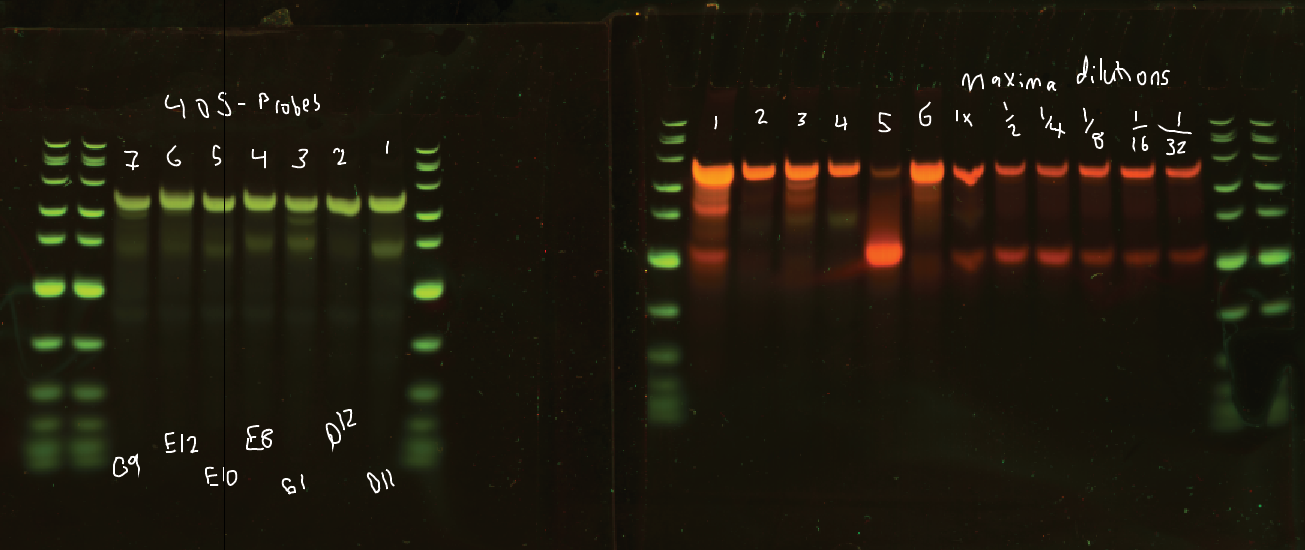
- Running denaturing gels.
- Ladder, Lanes 2-12 = cy3 RNA/primer ratios, probes 1-6, then Maxima 2x dilution-series enzyme titration.
- Gel 2 = 405 primers, probes 1-7.
Probe cleanup notes
- 1000 nmol of product = 40 ug DNA which is 8 fold greater than the reported 5 ug capacity of the Zymo CC-5 columns.
- RNA XP Agincourt Beads are supposed to bind DNA and RNA according to the specs. Apparently they don’t bind probe labeled DNA though.
- Probe cleanup: 405-labeled samples 1-3 cleanup using CC-25 columns (800 pmol)
- 405-labeled samples 4-6 cleanup using CC-5 columns
- samples 7-405 labeled and 1-cy3 labeled (1000 pmol scale) attempt to cleanup with beads. Failed completely — all the color stays in the solution. Elute from beads contains very little color (first wash in EtOH 700 uL contained substantial color. Left wash on too long (10 min?).
- Rescued 7-405 and cy3 by washing the non-bound fractions through cc-25 columns.
- Nanodrop K curve saturates at ~1.6
- Nanodrop for #6 cc5 elution: Red 0.4, Black = 1.67 (saturated). 5x dilution: Red = 0.08, Black = .692.
- Calc 0.69 * 5 *10/.027 = 1,282 ng/uL. = 32 pmol/uL. If full yield 800 pmol probe in 15 uL would be 52 pmol/uL.
- Second elute 0.14*10/.027 = 50 ng/uL = 1.25 pmol/uL
- Second round eluting flow-through 0.132*10/.027 ~ 50ng/uL ~ 1.25 pmol/uL. What happened to the other 20 pmol/uL ?!
Useful numbers
- Nanodrop absorption reading * 10 to go from mm^-1 to cm^-1 / 0.027 for ssDNA (0.02 for DNA).
Nanodrop UV-Vis results
- #1 D11-cy3 = .4 R, .514 Cy3 =
- #2 D12-cy3 = .3 R, .235 Cy3 =
- #3 E1-cy3 = .4 R, .38 Cy3 =
- #4 E8-cy3 = .27 R, .18 Cy3 =
- #5 E10-cy3 = .40 R, .87 Cy3 = (FAILED: all primer)
- #6 E12-cy3 = .60 R, .59 Cy3 =
- #1 D11-405 = ?1.3K ?.4 R, ~.025 405 = If full yield = 800 pmol in 30 uL = 26 pmol/uL.
- #2 D12-405 = ?1.9K ?.4 R, ~.018 405
- #3 E1-405 = ?1.6K ?.4 R,
- #4 E8-405 =
- #5 E10-405
- #6 E12-405
- #7 G9-405 = .2 R , 0.016 405 =
- second-round #4-405 = .09 K
- second-round #5-405 = .06 K
- second-round #6-405 = .132 K
Raw Nanodrop UV-Vis data
Ph Project
- very small difference over previous parameter range explored — most loci bound with small number of molecules
- Should compute radius of gyration and plot as a function of time to ensure convergence.
- Added radius of gyration calculation.
- small bug in treatment of bound-clusters: Need to consider “partner’s partner’s” — the bond needs to be transitive.
Mentoring
- New probes: F7, G1, F2, F4, F5.
- Student used wash buffer without EtOH. Need to set up separate bench with separate reagents to avoid confusion.