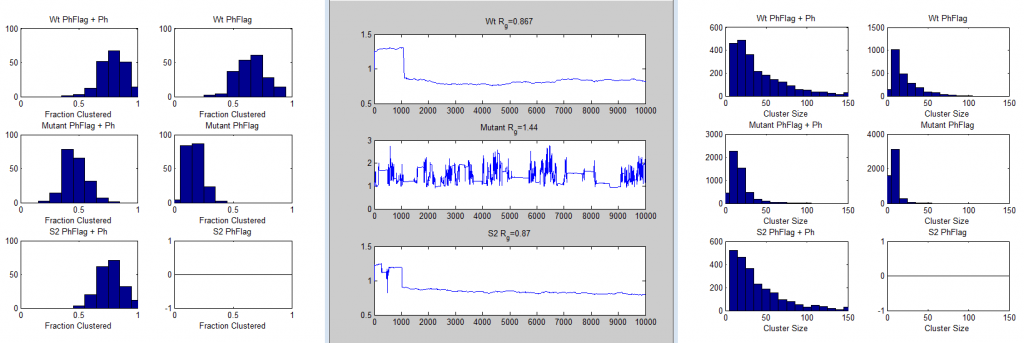
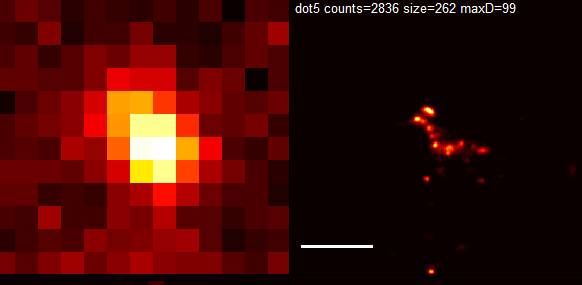
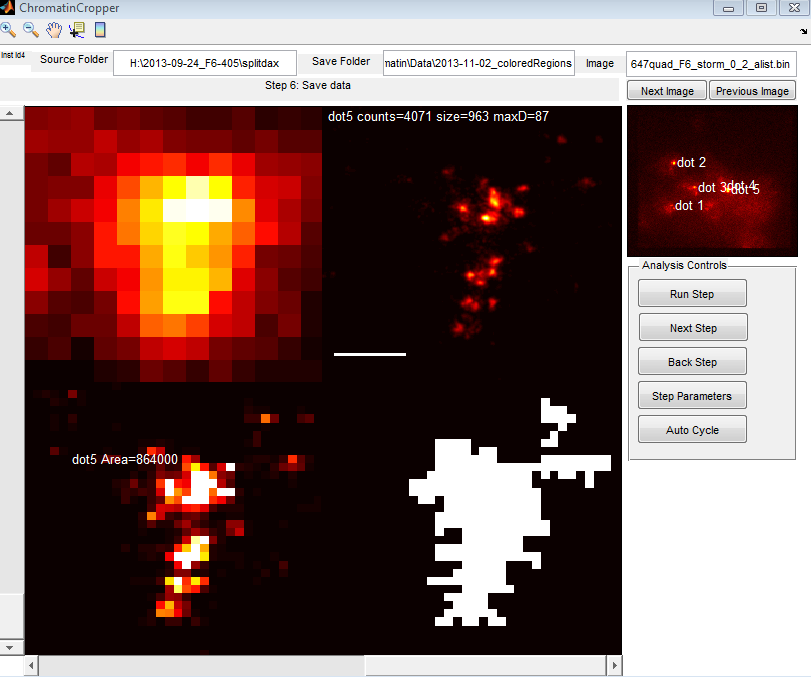
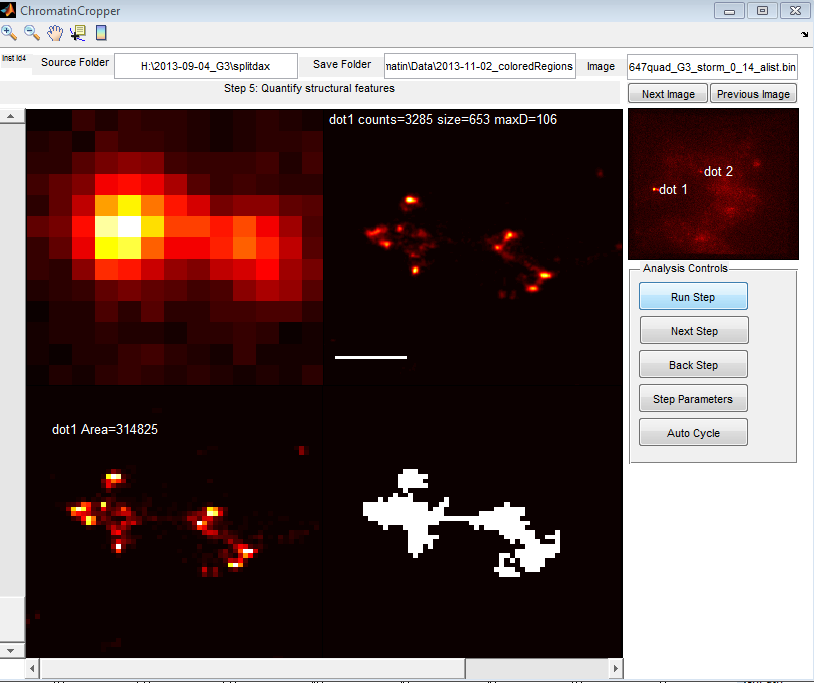
9:00a – 12:00p remotely.
12:00p – 12:00a
Goals
Coding
- Add to ChromatinCropper export
- labeled overview fig,
- object centroid and bounding box in original coordinates (for computation of distance to lamina)
- direct measure of distance to lamina
- New functionalities for ChromatinCropper
- needs an auto-cycle
Banff Report
- Finish writing this draft!
- send to co-organizers for edits
- Bibliography for workshop: list of publications of presented works
Ph Modeling
STORM Analysis
- Start new version of splitdax running on all unprocessed datasets.
- Determine fitting parameters for all unprocessed datasets.
D11
- D11 not well sealed? Bleaching? substantial progressive loss in localization density / brightness of individual switching. Further investigation, seems to be a spontaneous power loss of 647. Dyes much dimmer and on for much longer (5-40 frames instead of 1-2).
- maybe this is due to the bleaching during the conventional movie with max power 488.
- No beads taken with dataset D11.
- lamina bleached out, not good for a green region where we want to make this distance measurement. Of course we will need this distance measurement for yellow/blue/red as well to make comparisons.
Analyzing data
- BX-C s3
- Y F6. observation: Moment of interia should be corrected for mass. Y and BX-C have similar moments of inertia but a lot of the blue spots have a lot more mass.
- Analyze all non-K data through ProBox1. Saved in
2013-11-02_coloredRegions,Plot_ChromatinDots_131102.m
Cell Culture
- Kc cultures appear to be contaminated. Media is definitely contaminated.
- Made up new media yesterday.
- Start up new culture from frozen stock.
- Need to give BB his own media, cells, and supplies, so we can avoid cross-contamination.
- add P/S to working vial of media (stock is 10,000 units/mL, working conc. is 50 units/mL).